大豆(Glycine max)籽粒中蛋白质含量约40%,为人类提供了约30%的植物蛋白[1]。在我国,基本形成了进口大豆用于榨油、国产大豆用于蛋白食品消费的产业格局。培育高蛋白大豆品种,是满足未来大豆蛋白需求的根本途径[2]。然而,由于大豆蛋白含量是受多基因控制的复杂数量性状,且与产量负相关,依靠常规育种技术培育高蛋白、高产品种难度大[3]。“十二五”以来,东北、黄淮两大大豆主产区少有高蛋白品种(蛋白含量大于45%)通过国家区域试验审定。
野生大豆是提高栽培大豆蛋白质含量的优异基因源[4]。国家农作物种质保存中心保存一年生野生大豆种质资源6 000余份,蛋白质含量较栽培大豆高出5百分点,最高可达到55.4%[5-7]。野生大豆遗传多样性显著高于栽培大豆。一年生野生大豆驯化为栽培大豆地方品种,基因多样性降低50%,稀有等位变异丢失80%,等位基因频率发生显著性改变的基因比例达到60%。现代育成品种与一年生野生大豆相比,核苷酸多态性θ值、π值和单倍性多样性分别降低65%,49%,44%[8]。而且,聚类分析可将栽培大豆和一年生野生大豆分为独立的两组,表明栽培大豆和野生大豆间出现遗传分化[9]。王金陵等[10]从栽培大豆×野生大豆组合后代中获得6个蛋白质含量大于50%的株系;杨光宇等[11]选育出4个蛋白质含量超过50%且产量水平较理想的直立或半直立型株系。
受表型鉴定和基因型鉴定能力的制约,挖掘野生大豆高蛋白优异基因的理论工作滞后于育种实践。在表型鉴定方面,由于野生大豆具有蔓生、匍匐、缠绕等众多不利性状,表型鉴定十分繁琐,费时费力。而且野生大豆籽粒多为黑色,近红外分析不易获得准确地蛋白质含量检测结果,必须依赖于凯氏定氮法检测蛋白质含量[12]。在基因型鉴定方面,特别是大豆基因组测序结果公布之前,基因型检测费用昂贵,因此未见大量野生大豆关联分析报道。目前仅报道3个野生大豆高蛋白QTL,由Diers等[13]在A81-356022×PI468916组合中定位获得。第1个位于15号染色体,与SSR标记Satt411连锁;第2个位于13号染色体,与SSR标记Satt510和Satt335连锁;第3个位于20号染色体,与SSR标记Satt587和Satt239连锁。
随着生物信息技术的发展,美国农业部种质资源信息网(Germplasm Resources Information Network,GRIN)公布了一千余份野生大豆蛋白质含量检测数据(http://www.ars-grin.gov/npgs/searchgrin.html),同时,Song等[14]公布了上述野生大豆种质资源的SoySNP50K基因芯片数据。本研究旨在利用公共数据库中的表型和基因型数据,通过全基因组关联分析,挖掘与野生大豆高蛋白优异基因紧密连锁的SNP标记,为下一步通过回交将野生大豆高蛋白基因导入栽培大豆育成品种奠定基础,一方面筛选确定用于回交的野生亲本,同时开发可用于辅助选择的分子标记。
1 材料和方法
1.1 参试材料
用于本研究的野生大豆资源共508份,涵盖第4,5,6,7,8熟期组,分别从中国、韩国和日本采集得到[15]。
1.2 基因型和表型数据获取
参试508份野生大豆蛋白含量数据从美国农业部种质资源信息网下载(http://www.ars-grin.gov/npgs/searchgrin.html)。种子收获晾干后,利用凯氏定氮法测量大豆籽粒蛋白质含量。
利用Illumina公司大豆50K芯片(SoySNP50K BeadChip)检测参试材料SNP位点基因型。该芯片中包含位于大豆基因组常染色质区和异染色质区的52 041个SNP位点[14]。SNP基因型数据从SOYBASE网站下载(https://www.soybase.org/)。删除低频率等位变异小于5%的位点进行SNP基因型质量控制[16]。
1.3 群体遗传结构划分
根据连锁不平衡区段分析,共筛选出7 244个位于不同连锁不平衡区段的SNP位点,进行遗传结构分析。应用STRUCTURE 2.3.4中混合模型和等位变异发生频率非相关模型分析参试材料遗传结构[17]。所设置的STRUCTURE分析群体数(K)为2~7,每个K运行2次。每次运行的burnin time 和重复次数都设置为100 000。LnP(D)和 Evanno’s Δk 2种参数用于确定分组K值,其中,Δk=M[|L(k-1)-2L(k)+L(k+1)|]/s[L(k)],L(k)表示第k个LnP(D),M是2次运算的平均值,s是标准差。
1.4 全基因组关联分析
利用TASSEL软件中的混合线性模型(Mixed Linear Model,MLM)进行全基因组关联分析[18]。将某个SNP的-Log(p)>3.0设置为显著性阈值。Kinship值由TASSEL软件估算。
2 结果与分析
2.1 参试材料蛋白含量分布特征
参试508份野生大豆蛋白含量呈正态分布,蛋白含量最小值为38.1%,最大值56.9%,平均48.1%,标准差2.71%(图1)。蛋白含量最集中的范围是48%~50%,共143份,约占参试总材料份数的28.1%,其次为46%~48%,共123份,约占参试总材料份数的24.2%。蛋白质含量低于40%的材料仅有1份,即PI424096,蛋白含量38.1%;蛋白含量超过54%的材料有4份,最高为PI407301,达到56.9%,其他3份材料依次是PI407246、PI407322和PI407232,蛋白含量分别为56.8%,54.6%,54.4%。
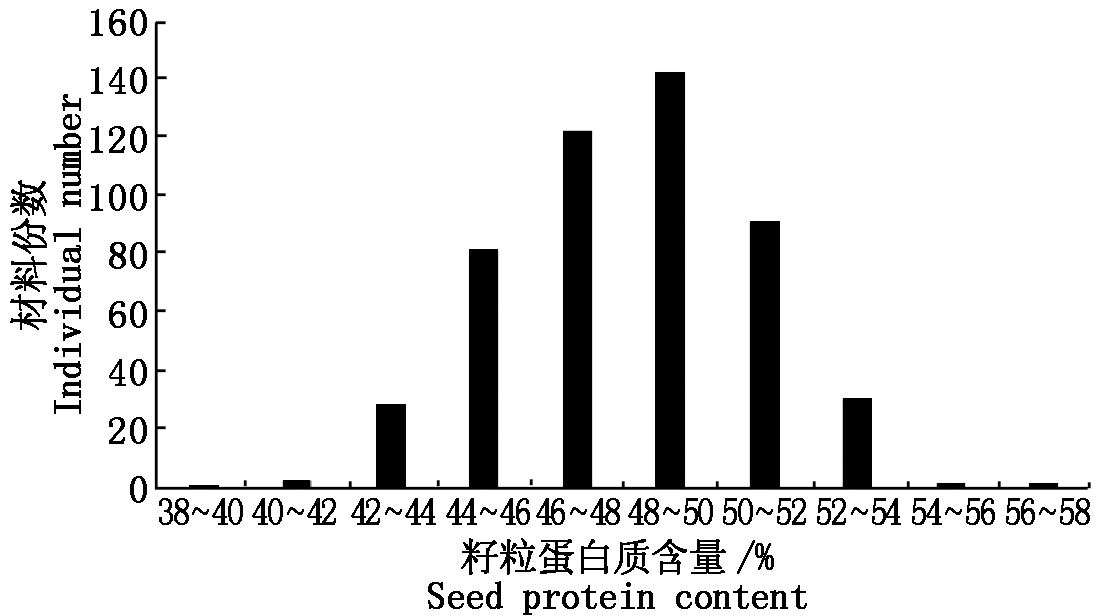
图1 参试野生大豆籽粒蛋白质含量分布
Fig.1 The distribution of protein content among the
wild soybean panel in this study
2.2 参试材料群体结构特征
对参试材料进行群体遗传结构分析,当分组数(K值)由2增加到7时,后验概率值(LnP(D))由-3 156 285升高到-3 013 257,但未出现明显拐点(图2-A)。当分3组时(K=3),ΔK获得最大值(图2-B),因此确定分组数为3(图2-C)。第1组中包括271份材料,蛋白含量最小值38.1%,最大值56.9%,平均为(48.3±2.92)%;第2组中包括111份材料,蛋白含量变化为42.0%~52.7%,平均(47.8±2.31)%;第3组中包括126份材料,蛋白含量42.5%~53.0%,平均(48.1±2.56)%。
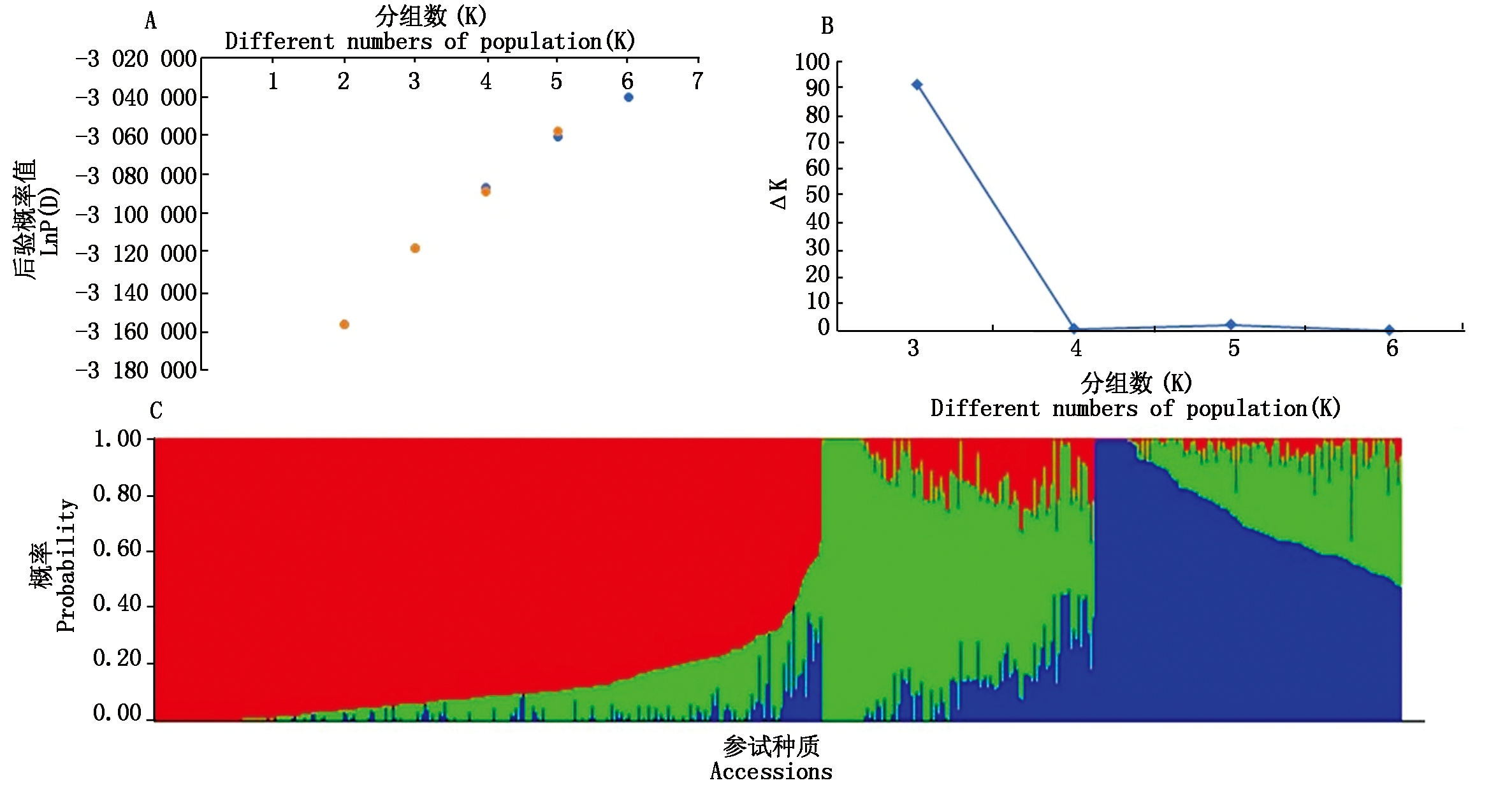
A.LnP(D)散点图;B.Δk散点图;C.群体结构划分图。
A.LnP(D)plot;B.Δk plot;C.The population structure.
图2 参试材料基于后验概率的群体遗传结构划分
Fig.2 The population structure analysis among the wild soybean panel
2.3 蛋白含量全基因组关联分析
为了对SNP位点进行质量控制,删除低频率等位变异频率小于0.05的SNP后,保留32 049个SNP用于全基因组关联分析。QQ散点图(Quantile-quantile plots)显示(图3),LOD实际观测值,与无关联位点预期值无明显背离,表明关联分析结果中假阳性比例在可接受范围内。通过全基因组关联分析,共检测到与蛋白含量相关的SNP位点74个,这些位点散布在19条染色体的60个单倍型区段内(表1)。显著性SNP位点LOD平均值为3.47,第1号染色体上的SNP位点BARC_1.01_Gm_01_54656209_A_G的LOD值最大,为5.18。单个显著性SNP位点可解释遗传变异为0.022~0.047,平均为0.027%。可解释遗传变异最大的SNP位点是第19号染色体上的BARC_1.01_Gm_19_36780878_G_T;第17号染色体上的BARC_1.01_Gm_17_7641259_C_T,可解释遗传变异最小。
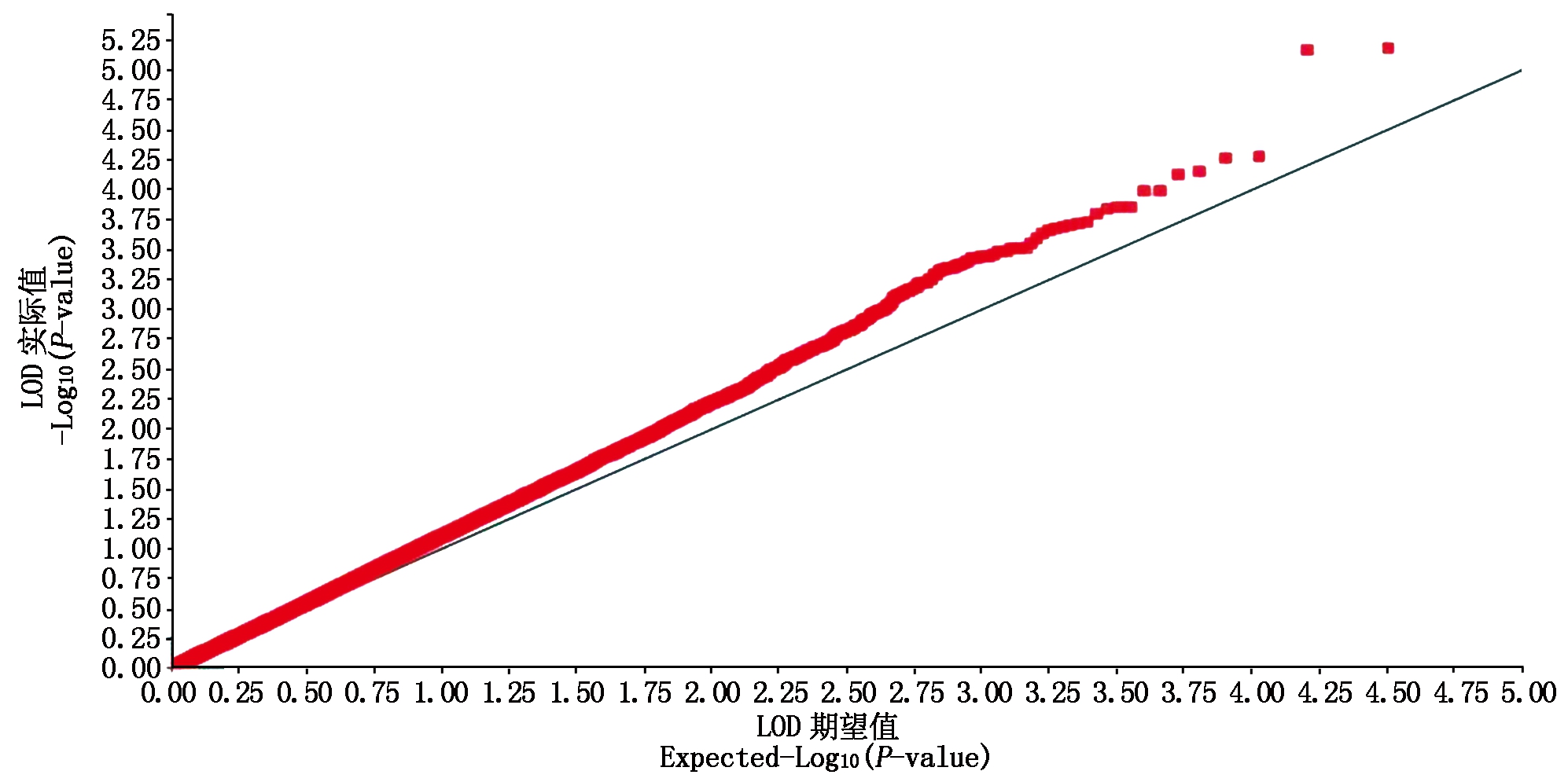
图3 蛋白含量混合线性模型QQ散点图
Fig.3 Quantile-quantile plots of the mixed linear model(MLM)for seed protein
表1 在野生大豆中检测到的与蛋白含量相关的SNP
Tab.1 SNPs associated with protein content by MLM in the panel Soja_MS_9899
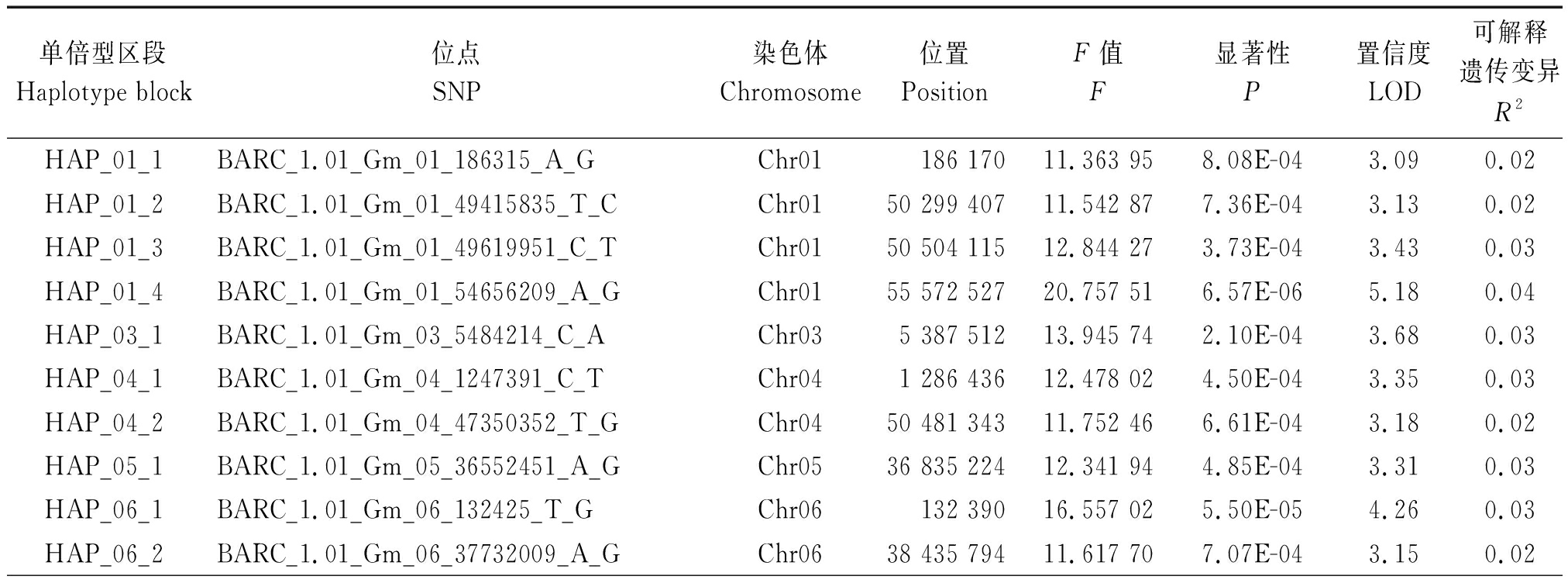
单倍型区段Haplotype block位点SNP染色体Chromosome位置PositionF值F显著性P置信度LOD可解释遗传变异R2HAP_01_1BARC_1.01_Gm_01_186315_A_GChr01186 17011.363 958.08E-043.09 0.02 HAP_01_2BARC_1.01_Gm_01_49415835_T_CChr0150 299 40711.542 877.36E-043.13 0.02 HAP_01_3BARC_1.01_Gm_01_49619951_C_TChr0150 504 11512.844 273.73E-043.43 0.03 HAP_01_4BARC_1.01_Gm_01_54656209_A_GChr0155 572 52720.757 516.57E-065.18 0.04 HAP_03_1BARC_1.01_Gm_03_5484214_C_AChr035 387 51213.945 742.10E-043.68 0.03 HAP_04_1BARC_1.01_Gm_04_1247391_C_TChr041 286 43612.478 024.50E-043.35 0.03 HAP_04_2BARC_1.01_Gm_04_47350352_T_GChr0450 481 34311.752 466.61E-043.18 0.02 HAP_05_1BARC_1.01_Gm_05_36552451_A_GChr0536 835 22412.341 944.85E-043.31 0.03 HAP_06_1BARC_1.01_Gm_06_132425_T_GChr06132 39016.557 025.50E-054.26 0.03 HAP_06_2BARC_1.01_Gm_06_37732009_A_GChr0638 435 79411.617 707.07E-043.15 0.02
表1(续)

单倍型区段Haplotype block位点SNP染色体Chromosome位置PositionF值F显著性P置信度LOD可解释遗传变异R2HAP_06_3BARC_1.01_Gm_06_47586108_C_TChr0648 116 22913.052 943.34E-043.48 0.03 HAP_06_4BARC_1.01_Gm_06_48998713_C_TChr0649 599 02913.169 543.14E-043.50 0.03 HAP_07_1BARC_1.01_Gm_07_649600_T_CChr07654 63312.251 245.07E-043.29 0.02 HAP_07_2BARC_1.01_Gm_07_17637298_A_GChr0717 721 31212.064 565.60E-043.25 0.03 HAP_07_3BARC_1.01_Gm_07_37394702_C_TChr0737 300 29911.481 007.66E-043.12 0.03 HAP_08_1BARC_1.01_Gm_08_9885021_T_CChr089 877 09813.196 033.11E-043.51 0.03 HAP_08_2BARC_1.01_Gm_08_20979500_T_CChr0820 936 83512.208 385.22E-043.28 0.03 HAP_08_3BARC_1.01_Gm_08_21133515_G_AChr0821 090 26412.376 664.74E-043.32 0.03 HAP_08_4BARC_1.01_Gm_08_29906080_T_CChr0830 555 32511.910 466.06E-043.22 0.02 HAP_08_5BARC_1.01_Gm_08_35742525_T_CChr0836 351 01613.370 192.86E-043.54 0.03 HAP_08_6BARC_1.01_Gm_08_43206458_A_CChr0844 041 43314.745 821.39E-043.86 0.03 HAP_08_7BARC_1.01_Gm_08_44448158_C_AChr0845 323 76211.516 037.48E-043.13 0.02 HAP_09_1BARC_1.01_Gm_09_3708901_T_CChr093 751 12410.963 721.00E-033.00 0.02 HAP_10_1BARC_1.01_Gm_10_5477657_G_AChr105 509 48612.919 703.58E-043.45 0.03 HAP_10_2BARC_1.01_Gm_10_48281152_G_AChr1048 876 84114.731 441.40E-043.85 0.03 HAP_10_3BARC_1.01_Gm_10_50095630_G_TChr1050 692 84013.179 543.13E-043.50 0.03 HAP_11_1BARC_1.01_Gm_11_15151298_C_TChr1124 655 82911.401 007.92E-043.10 0.02 BARC_1.01_Gm_11_15154068_G_AChr1124 658 59911.479 887.60E-043.12 0.02 BARC_1.01_Gm_11_15162117_G_AChr1124 666 64811.602 277.13E-043.15 0.02 BARC_1.01_Gm_11_15167305_G_AChr1124 671 83614.466 761.60E-043.80 0.03 BARC_1.01_Gm_11_15167362_C_AChr1124 671 89312.846 563.71E-043.43 0.03 BARC_1.01_Gm_11_15168038_C_AChr1124 672 56913.986 202.06E-043.69 0.03 BARC_1.01_Gm_11_15168696_C_TChr1124 673 22712.518 074.41E-043.36 0.03 BARC_1.01_Gm_11_15171742_A_CChr1124 676 27311.701 906.77E-043.17 0.02 BARC_1.01_Gm_11_15215584_C_TChr1124 720 11511.203 048.79E-043.06 0.02 HAP_11_2BARC_1.01_Gm_11_2211407_C_TChr112 219 78414.669 601.45E-043.84 0.03 BARC_1.01_Gm_11_2211528_G_TChr112 219 90514.058 091.99E-043.70 0.03 HAP_11_3BARC_1.01_Gm_11_37237162_G_TChr1132 785 13012.433 254.63E-043.33 0.03 HAP_12_1BARC_1.01_Gm_12_27500547_T_CChr1223 870 23514.150 791.90E-043.72 0.03 BARC_1.01_Gm_12_27563620_C_TChr1223 933 30815.922 947.62E-054.12 0.03 BARC_1.01_Gm_12_27646431_A_GChr1224 015 91115.356 391.03E-043.99 0.03 BARC_1.01_Gm_12_27678060_A_GChr1224 052 70011.101 439.31E-043.03 0.02 HAP_12_2BARC_1.01_Gm_12_7182847_G_AChr127 221 51212.033 055.68E-043.25 0.02 HAP_12_3BARC_1.01_Gm_12_25979150_T_CChr1222 183 00912.867 223.71E-043.43 0.03 HAP_13_1BARC_1.01_Gm_13_30221939_C_TChr1331 403 11913.046 613.35E-043.47 0.03 HAP_13_2BARC_1.01_Gm_13_40531608_G_AChr1341 977 20112.473 504.54E-043.34 0.03 HAP_14_1BARC_1.01_Gm_14_6354474_T_CChr146 460 92716.616 955.33E-054.27 0.03 HAP_14_2BARC_1.01_Gm_14_8755748_C_TChr148 547 22314.122 911.92E-043.72 0.03 HAP_14_3BARC_1.01_Gm_14_13385608_T_CChr1413 187 21813.231 293.09E-043.51 0.03 HAP_14_4BARC_1.01_Gm_14_20221695_A_CChr1424 367 69813.106 853.28E-043.48 0.03 HAP_15_1BARC_1.01_Gm_15_578068_A_CChr15577 71311.947 915.94E-043.23 0.02 HAP_15_2BARC_1.01_Gm_15_3614684_T_CChr153 632 90013.733 682.35E-043.63 0.03 HAP_15_3BARC_1.01_Gm_15_4775310_C_TChr154 794 79112.633 564.16E-043.38 0.03 HAP_15_4BARC_1.01_Gm_15_9012474_T_CChr159 054 53820.663 746.90E-065.16 0.04 HAP_15_5BARC_1.01_Gm_15_10537858_A_GChr1510 614 91211.144 969.07E-043.04 0.02 HAP_15_6BARC_1.01_Gm_15_30127024_C_TChr1532 629 63211.863 096.21E-043.21 0.02 HAP_15_7BARC_1.01_Gm_15_41081388_C_TChr1541 824 55611.674 416.87E-043.16 0.02 HAP_16_1BARC_1.01_Gm_16_4936997_G_AChr164 965 24012.920 443.58E-043.45 0.03
表1(续)
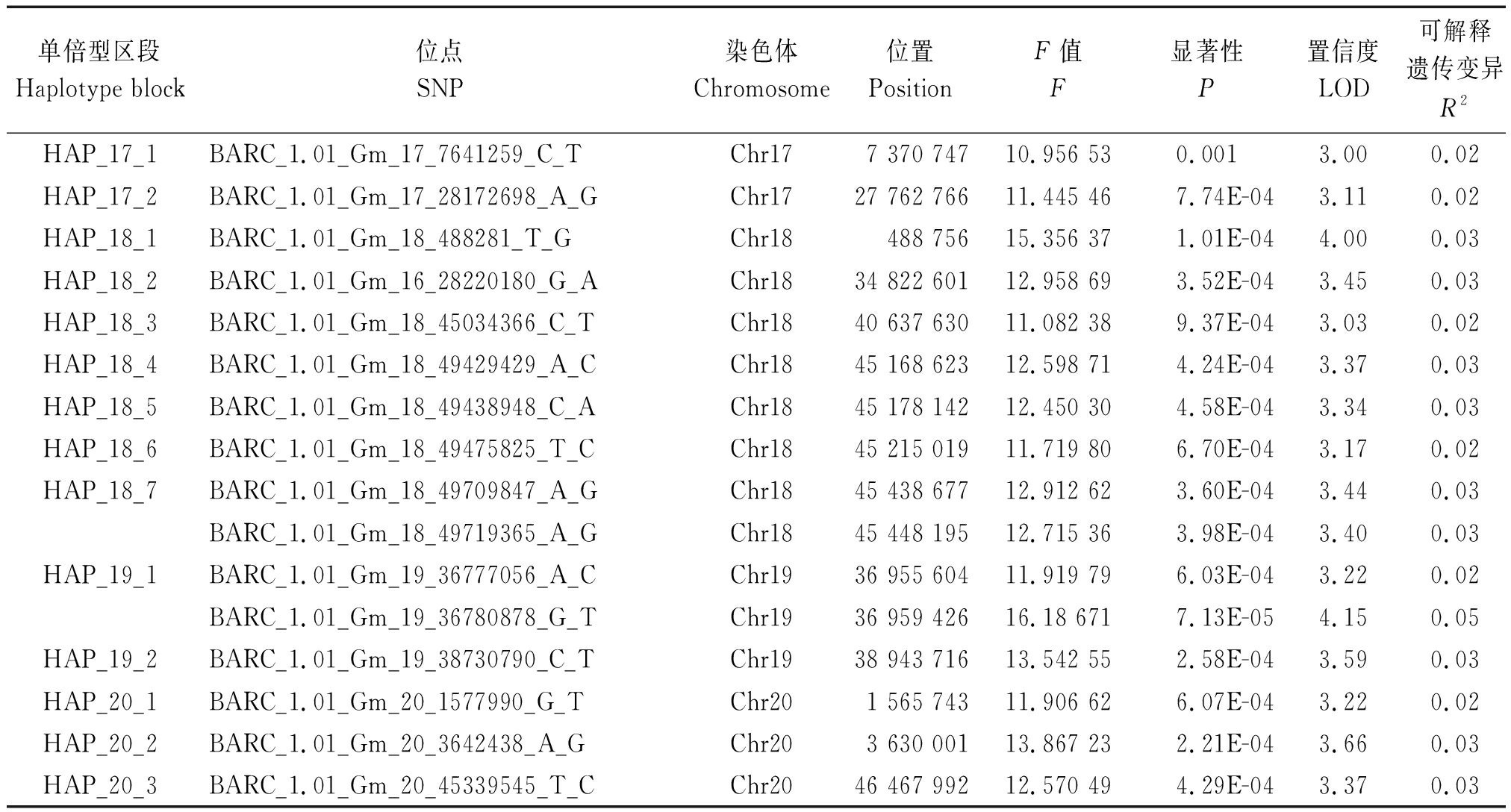
单倍型区段Haplotype block位点SNP染色体Chromosome位置PositionF值F显著性P置信度LOD可解释遗传变异R2HAP_17_1BARC_1.01_Gm_17_7641259_C_TChr177 370 74710.956 530.0013.00 0.02 HAP_17_2BARC_1.01_Gm_17_28172698_A_GChr1727 762 76611.445 467.74E-043.11 0.02 HAP_18_1BARC_1.01_Gm_18_488281_T_GChr18488 75615.356 371.01E-044.00 0.03 HAP_18_2BARC_1.01_Gm_16_28220180_G_AChr1834 822 60112.958 693.52E-043.45 0.03 HAP_18_3BARC_1.01_Gm_18_45034366_C_TChr1840 637 63011.082 389.37E-043.03 0.02 HAP_18_4BARC_1.01_Gm_18_49429429_A_CChr1845 168 62312.598 714.24E-043.37 0.03 HAP_18_5BARC_1.01_Gm_18_49438948_C_AChr1845 178 14212.450 304.58E-043.34 0.03 HAP_18_6BARC_1.01_Gm_18_49475825_T_CChr1845 215 01911.719 806.70E-043.17 0.02 HAP_18_7BARC_1.01_Gm_18_49709847_A_GChr1845 438 67712.912 623.60E-043.44 0.03 BARC_1.01_Gm_18_49719365_A_GChr1845 448 19512.715 363.98E-043.40 0.03 HAP_19_1BARC_1.01_Gm_19_36777056_A_CChr1936 955 60411.919 796.03E-043.22 0.02 BARC_1.01_Gm_19_36780878_G_TChr1936 959 42616.18 6717.13E-054.15 0.05 HAP_19_2BARC_1.01_Gm_19_38730790_C_TChr1938 943 71613.542 552.58E-043.59 0.03 HAP_20_1BARC_1.01_Gm_20_1577990_G_TChr201 565 74311.906 626.07E-043.22 0.02 HAP_20_2BARC_1.01_Gm_20_3642438_A_GChr203 630 00113.867 232.21E-043.66 0.03 HAP_20_3BARC_1.01_Gm_20_45339545_T_CChr2046 467 99212.570 494.29E-043.37 0.03
全部74个显著性SNP位点中,9个富集在第11号染色体常染色质区15 128 832~15 253 199,长度为124 kb的单倍型区段内,文中命名为HAP_11_1。另外4个显著性SNP富集于第12号染色体异染色质区26 842 687~27 818 244,长度为976 kb的单倍型区段内,文中命名为HAP_12_1。在HAP_11_1中,经SNP质量控制后,用于本研究关联分析的SNP位点共30个,LOD值变化为0.01~3.80,平均为1.39,可解释遗传变异变化为0~2.88%,平均为1.00%;其中,9个显著性SNP位点LOD值平均为3.32,可解释遗传变异平均为2.50%;SNP位点BARC_1.01_Gm_11_15167305_G_A的LOD值最大,为3.80,可解释遗传变异为2.88%。在HAP_12_1中,经SNP质量控制后,用于关联分析的SNP位点共9个,LOD值变化介于0.41~4.12,平均为2.27,可解释遗传变异变化在0~3.36%,平均为1.71%;其中,4个显著性SNP位点LOD值平均为3.71,可解释遗传变异平均为2.98%;SNP位点BARC_1.01_Gm_12_27563620_C_T的LOD值最大,为4.12,可解释遗传变异为3.23%。
3 讨论与结论
本研究中发现2个与蛋白含量相关的单倍型区段,HAP_11_1和HAP_12_1。前人利用双亲杂交分离群体在相近位置处发现了与蛋白含量相关的分子标记。在科丰1号×南农1138-2组合中,检测到RFLP标记A520_1与蛋白含量连锁[19],参照Soymap3 [20],及大豆物理位置[21],与HAP_11_1位置重叠,A520_1可解释总遗传变异的5.8%。在豫豆12×ZDD09454组合中,检测到SSR标记Satt469与蛋白含量连锁[22],与HAP_12_1位置重叠。在OAC Wallace×OAC Glencoe组合中,Satt302与蛋白含量连锁[23],与HAP_12_1位置重叠。但Hwang等[24]利用300份栽培大豆为材料,进行关联分析时,没有检测到这2个区间与蛋白含量的相关性。这2个区间与蛋白含量的相关性,在本研究中利用野生大豆自然作图群体可检测到,文献报道在部分双亲分离群体中可检测到,而在栽培大豆自然群体中没有检测到。可能原因是这2个区间里调控蛋白含量的基因,在野生大豆中,不同等位变异分布较均匀,而在驯化过程中,部分等位变异被淘汰,成为稀有等位变异,所以能够在特定的双亲分离群体中检测到,而不能在自然群体中检测到[25],因此挖掘野生大豆优异基因,有可能进一步扩大栽培大豆遗传基础[26]。
大豆基因组中存在大量不同长度、包含不同数量的SNP的单倍型区间[16]。同一单倍型区间内的不同SNP,通常作为一个整体出现,不易发生重组。因此,同一单倍型区间内的不同SNP,应该与特定的性状具有相对一致的相关性。但在本研究中,HAP_11_1和HAP_12_1,均有部分SNP与蛋白含量相关性达不到显著性水平。经分析,次要等位变异基因频率与LOD值呈正相关关系,相关系数81.2%。在次要等位变异频率大于25%的20个SNP中,仅有3个LOD值小于2.5,分别是BARC_1.01_Gm_11_15215338_C_T、BARC_1.01_Gm_12_27170294_C_A和BARC_1.01_Gm_12_27818244_A_G。其中,BARC_1.01_Gm_12_27170294_C_A在分3组后,每组内次要等位变异频率均小于25%。因此,以选择基因型群体为作图群体,可能更容易发觉到重要的稀有等位变异[27]。
这2个区间在驯化和育种选择过程中,经受了不同的选择压力。其中,HAP_11_1区间,野生大豆与地方品种之间的分化系数Fst平均值为0.042,地方品种和育成品种间的分化系数Fst平均值为0.151;HAP_12_1区间,野生大豆与地方品种之间的分化系数Fst平均值为0.759,地方品种和育成品种间的分化系数Fst平均值为0.001 [16]。表明驯化过程造成HAP_12_1区间基因频率显著性改变。由此推论,在挖掘利用这2个区间的蛋白含量优异基因过程中,针对HAP_11_1区间,可以首先尝试在育成品种和地方品种里寻找优异基因;而针对HAP_12_1区间,则有必要回到野生资源里寻找优异基因。其他的品质相关性状经受驯化、选择压力也有类似报道[28]
[1] 杨春燕,姚利波,刘兵强,张孟臣.国内外大豆品质育种研究方法与最新进展[J].华北农学报,2009,24(S1):75-78.doi:10.7668/hbnxb.2009.S1.019.
Yang C Y,Yao L B,Liu B Q,Zhang M C.Advance on soybean quality breeding in China and abroad[J].Acta Agriculturae Boreali-Sinica,2009,24(S1):75-78.
[2] 盖钧镒.大豆加工业的发展及其对大豆品质的要求[J].农产品加工,2008(7):4-7.doi:10.3969/j.issn.1671-9646-C.2008.07.002.
Gai J Y.Development of soybean processing industry and its requirements for soybean quality[J].Farm Products processing,2008(7):4-7.
[3] Rincker K,Nelson R,Specht J,Sleper D,Cary T,Cianzio S R,Casteel S,Conley S,Chen P Y,Davis V,Fox C,Graef G,Godsey C,Holshouser D,Jiang G L,Kantartzi S K,Kenworthy W,Lee C,Mian R,McHale L,Naeve S,Orf J,Poysa V,Schapaugh W,Shannon G,Uniatowski R,Wang D C,Diers B.Genetic improvement of US soybean in maturity groups Ⅱ,Ⅲ,and Ⅳ[J].Crop Science,2014,54(4):1419-1432.doi:10.2135/cropsci2013.10.0665.
[4] Yan L,Xing L L,Yang C Y,Chang R Z,Zhang M C,Qiu L J.Identification of quantitative trait loci associated with soybean seed protein content using two populations derived from crosses between Glycine max and Glycine soja[J].Plant Genetic Resources,2014,12(S1):S104-S108.doi:10.1017/s1479262114000379.
[5] 中国农业科学院油料所.中国大豆品种资源目录[M].北京:农业出版社,1982.
Oil Crop Research Institute,Chinese Academy of Agricultural Sciences.Catalogues of chinese soybean germplasm resources[M].Beijing:China Agriculture Press,1982.
[6] 中国农业科学院作物品种资源所.中国大豆品种资源目录(续编一)[M].北京:农业出版社,1991.
Institute of Crop Germplasm Resources,Chinese Academy of Agricultural Sciences.Catalogues of Chinese soybean germplasm resources(Continuation Ⅰ)[M].Beijing:China Agriculture Press,1991.
[7] 中国农业科学院作物品种资源所.中国大豆品种资源目录(续编二)[M].北京:中国农业出版社,1996.
Institute of Crop Germplasm Resources,Chinese Academy of Agricultural Sciences.Catalogues of Chinese soybean germplasm resources(Continuation Ⅱ)[M].Beijing:China Agriculture Press,1996.
[8] Hyten D L,Song Q J,Zhu Y L,Choi I Y,Nelson R L,Costa J M,Specht J E,Shoemaker R C,Cregan P B.Impacts of genetic bottlenecks on soybean genome diversity[J].PNAS,2006,103(45):16666-16671.doi:10.1073/pnas.0604379103.
[9] Li Y H,Guan R X,Liu Z X,Ma Y S,Wang L X,Li L H,et al.Genetic structure and diversity of cultivated soybean(Glycine max (L.)Merr.)landraces in China[J].Theoretical and Applied Genetics,2008,117(6):857-871.doi:10.1007/s00122-008-0825-0.
[10] 王金陵,孟庆喜,杨庆凯,赵淑文,武天龙.回交对克服栽培大豆与野生和半野生大豆杂交后代蔓生倒伏性的效应[J].大豆科学,1986,5(3):181-187.doi:10.11861/j.issn.1000-9841.1986.03.0181.
Wang J L,Meng Q X,Yang Q K,Zhao S W,Wu T L.Effect OF backcrossing ON overcoming viny and lodging HABIt OF cultivated X wild and CULTIVATEDxSEMI-WILD crosses[J].Soybean Science,1986,5(3):181-187.
[11] 杨光宇,纪锋.中国野生大豆资源的研究与利用综述 Ⅰ.地理分布、化学品质性状及在育种中的利用[J].吉林农业科学,1999,24(1):12-17.doi:10.16423/j.cnki.1003-8701.1999.01.004.
Yang G Y,Ji F.Review on research and utilization of wild soybean resources in China Ⅰ.Geographical distribution,chemical quality traits and their utilization in breeding[J].Jilin Agricultural Sciences,1999,24(1):12-17.
[12] 闫龙,蒋春志,于向鸿,杨春燕,张孟臣.大豆粗蛋白、粗脂肪含量近红外检测模型建立及可靠性分析[J].大豆科学,2008,27(5):833-837.doi:10.11861/j.issn.1000-9841.2008.05.0833.
Yan L,Jiang C Z,Yu X H,Yang C Y,Zhang M C.Development and reliability of near infrared spectroscopy(NIS)models of protein and oil content in soybean[J].Soybean Science,2008,27(5):833-837.
[13] Diers B W,Keim P,Fehr W R,Shoemaker R C.RFLP analysis of soybean seed protein and oil content[J].Theoretical and Applied Genetics,1992,83(5):608-612.doi:10.1007/BF00226905.
[14] Song Q J,Hyten D L,Jia G F,Quigley C V,Fickus E W,Nelson R L,Cregan P B.Development and evaluation of SoySNP50K,a high-density genotyping array for soybean[J].PLoS One,2013,8(1):e54985.doi:10.1371/journal.pone.0054985.
[15] Chen Y W,Nelson R L.Evaluation and classification of leaflet shape and size in wild soybean[J].Crop Science,2004,44(2):671-677.doi:10.2135/cropsci2004.6710.
[16] Song Q J,Hyten D L,Jia G F,Quigley C V,Fickus E W,Nelson R L,Cregan P B.Fingerprinting soybean germplasm and its utility in genomic research[J].G3 Genes|Genomes|Genetics,2015,5(10):1999-2006.doi:10.1534/g3.115.019000.
[17] Pritchard J K,Stephens M,Donnelly P.Inference of population structure using multilocus genotype data[J].Genetics,2000,155(2):945-959.doi:10.1093/genetics/155.2.945.
[18] Bradbury P J,Zhang Z,Kroon D E,Casstevens T M,Ramdoss Y,Buckler E S.TASSEL:software for association mapping of complex traits in diverse samples[J].Bioinformatics,2007,23(19):2633-2635.doi:10.1093/bioinformatics/btm308.
[19] Gai J Y,Wang Y J,Wu X L,Chen S Y.A comparative study on segregation analysis and QTL mapping of quantitative traits in plants-with a case in soybean[J].Frontiers of Agriculture in China,2007,1(1):1-7.doi:10.1007/s11703-007-0001-3.
[20] Song Q J,Jia G F,Zhu Y L,Grant D,Nelson R T,Hwang E Y,Hyten D L,Cregan P B.Abundance of SSR motifs and development of candidate polymorphic SSR markers(BARCSOYSSR_1.0)in soybean[J].Crop Science,2010,50(5):1950-1960.doi:10.2135/cropsci2009.10.0607.
[21] Schmutz J,Cannon S B,Schlueter J,Ma J X,Mitros T,Nelson W,et al.Genome sequence of the palaeopolyploid soybean[J].Nature,2010,463(7278):178-183.doi:10.1038/nature08670. doi:10.1038/nature08670.
[22] Lu W G, Wen Z X, Li H C, Yuan D H, Li J Y, Zhang H, Huang Z W, Cui S Y, Du W. Identification of the quantitative trait loci (QTL) underlying water soluble protein content in soybean[J].Theoretical and Applied Genetics,2013,126(2):425-433.doi:10.1007/s00122-012-1990-8.
[23] Eskandari M,Cober, ER,Rajcan I. Genetic control of soybean seed oil: II. QTL and genes that increase oil concentration without decreasing protein or with increased seed yield[J].Theoretical and Applied Genetics,2013,126(6)1677-1687. doi:10.1007/s00122-013-2083-z.
[24] Hwang E Y,Song Q J,Jia G F,Specht J E,Hyten D L,Costa J,Cregan P B.A genome-wide association study of seed protein and oil content in soybean[J].BMC Genomics,2014,15:1.doi:10.1186/1471-2164-15-1.
[25] Zhou Z K,Jiang Y,Wang Z,Gou Z H,Lyu J,Li W Y,et al.Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean[J].Nature Biotechnology,2015,33(4):408-414.doi:10.1038/nbt.3096.
[26] Li Y H,Zhou G Y,Ma J X,Jiang W K,Jin L G,Zhang Z H,et al.De novo assembly of soybean wild relatives for Pan-genome analysis of diversity and agronomic traits[J].Nature Biotechnology,2014,32(10):1045-1052.doi:10.1038/nbt.2979.
[27] Yan L,Hofmann N,Li S X,Ferreira M E,Song B H,Jiang G L,Ren S X,Quigley C,Fickus E,Cregan P,Song Q J.Identification of QTL with large effect on seed weight in a selective population of soybean with genome-wide association and fixation index analyses[J].BMC Genomics,2017,18(1):529.doi:10.1186/s12864-017-3922-0.
[28] Yan L,Di R,Wu C J,Liu Q,Wei Y,Hou W H,Zhao Q S,Liu B Q,Yang C Y,Song Q J,Zhang M C.Haplotype analysis of a major and stable QTL underlying soybean(Glycine max)seed oil content reveals footprint of artificial selection[J].Molecular Breeding,2019,39(4):57.doi:10.1007/s11032-019-0951-1.
