生长素作为最早发现的一类植物激素,参与调控植物多种生长发育过程,如根的发育[1]、顶端优势、向性反应和形态建成[2]等。生长素调控通路中关键蛋白主要包括生长素/吲哚乙酸蛋白(Aux/IAAs)、SCF复合系统和生长素响应因子(ARFs)[3]。生长素响应因子是能与生长素应答元件(AuxRE)TGTCTC序列特异结合,调节生长素反应基因的一类转录因子,最先在模式植物拟南芥中被鉴定出来[4],该家族共有23个基因[5]。其中ARF1/ARF2功能的缺失通过增加拟南芥中Aux/IAA基因的转录进而调控叶片衰老和开花时间[6];ARF3/ARF4主要调控侧生器官的结构发育[7],ARF3直接与细胞分裂素基因AtIPT5启动子结合,负调控AtIPT5的表达,介导生长素与细胞分裂素相互作用,影响新生芽的再生[8];ARF5通过调控AMP1的表达影响胚和维管组织的形成[9];arf6、arf8突变体雄蕊发育迟缓,而arf6/arf8双突变体则不能形成成熟的花,表明ARF6、ARF8共同参与花的形态建成[10],同时,ARF8还负调控果实的起始发育[11];ARF7/ARF19可直接激活下游基因LBD/ASLs的表达来影响侧根形成[12];ARF10/ARF16则是在miR160下游调控根冠发育及根的向地性[13]。
单子叶植物水稻ARF家族中有25个成员[14]。OsARF1是水稻中第一个被发现的ARF基因[15],它与胚芽鞘的向性有关;之后的研究表明,OsARF1对营养生长和种子发育至关重要[16];OsARF4能与OsGSK41/OsGSK互作且被后者磷酸化,调控水稻籽粒大小及千粒质量[17];OsARF16参与细胞分裂素介导的水稻磷酸盐转运和信号传递通路,且敲除株系对外源细胞分裂素不响应[18]。OsARF17和OsARF19通过调节生长素和BR信号来控制水稻叶夹角大小[19-20]。然而,目前对水稻ARF家族的研究大多集中在籽粒发育和叶夹角等方面,ARFs其他成员对水稻农艺性状的影响尚不明确。
CRISPR/Cas9是第3代基因编辑技术,相比其他编辑技术具有成本低、快速高效等优点,已经成为目前最主流的基因编辑系统[21]。近年来,大量研究人员通过CRISPR/Cas9技术对水稻基因编辑,研究水稻基因的功能,也取得了重要进展[22]。本研究利用CRISPR/Cas9系统对水稻生长素响应因子OsARF12第1个和第2个外显子靶位点进行编辑,独立转化粳稻品种日本晴,通过对2个外显子不同突变位点的KO-ARF12-1和KO-ARF12-2突变体进行测序、表达量鉴定、表型性状调查,研究OsARF12对水稻农艺性状的影响。
1 材料和方法
1.1 试验材料
转基因受体材料为:粳稻品种日本晴(Oryza sativa spp. japonica cv. Nipponbare)。CRISPR/Cas9载体为:pOs-gRNA、pH-Ubi-CAS9。试验所需引物(表1)合成与测序分析均由上海生工生物工程股份有限公司完成。
表1 试验所用引物
Tab.1 Primers used in this test

引物名称Primer name引物序列(5′-3′)Primer sequence(5′-3′)ARF12 Crispr1_FGGCGGAACGATGCGTACCTTCCCGARF12 Crispr1_RAAACCGGGAAGGTACGCATCGTTCARF12 Crispr2_FGGCGTTCACACCCGGATATCGGTTARF12 Crispr2_RAAACAACCGATATCCGGGTGTGAACrispr ARF12-1_FCTGACTTGGGCTGGCAATTTCrispr ARF12-1_RGAGGCTGCTGTGTGAAATCCCrispr ARF12-2_FTTTCCTCCCCATGTACACCGCrispr ARF12-2_RATAATGGGGTGATGTCCTGACOsARF12_F定CTGACTTGGGCTGGCAATTTOsARF12_R定TCGACGAATACAAGCTGCCAGCactin_FGGAAGTACAGTGTCTGGATTGGAGactin_RTCTTGGCTTAGCATTCTTGGGT
1.2 试验方法
1.2.1 OsARF12靶位点设计和表达载体构建 根据CRISPR/Cas9原理,在RAP-DB网站(https://rapdb.dna.affrc.go.jp/)上获取水稻OsARF12外显子序列,分别选取第1个外显子PAM序列(CGG)前20 bp(5′-GAACGATGCGTACCTTCCCG-3′)及第2个外显子PAM序列(GGG)前20 bp(5′-TTCACACCCGGATATCGGTT-3′)为靶位点(图1)。分别在2个靶位点5′端前加上Bsa Ⅰ限制性内切酶的黏性末端接头GGCG,即为ARF12 Crispr1_F、ARF12 Crispr2_F(表1);将选取的2个靶序列分别反向互补并在其5′端前加上Bsa Ⅰ限制性内切酶的黏性末端接头AAAC,即为ARF12 Crispr1_R、ARF12 Crispr2_R(表1)。将2对加过接头的序列送往上海生工生物工程股份有限公司合成后,经磷酸化修饰和退火形成双链,用T4 DNA连接酶与经过限制性内切酶Bsa Ⅰ酶切过后的中间载体sgRNA连接,转化大肠杆菌感受态DH5α,随后进行菌落PCR检测与测序,并用阳性质粒与Cas9终载体进行LR重组,转化DH5α,经菌落PCR和测序检测,将阳性质粒转化农杆菌EHA105。利用农杆菌介导法导入粳稻品种日本晴,获得转基因植株。
1.2.2 植株DNA和总RNA的提取 在野生型和各转基因植株抽穗前取叶片样品,并立即置于液氮中,-80 ℃冰箱保存。采用CTAB法提取叶片DNA;TRIzol法提取总RNA。
1.2.3 反转录 使用天根生化科技有限公司FastKing RT Kit(KR118-02)反转录试剂盒将1 μg总RNA反转录成cDNA。
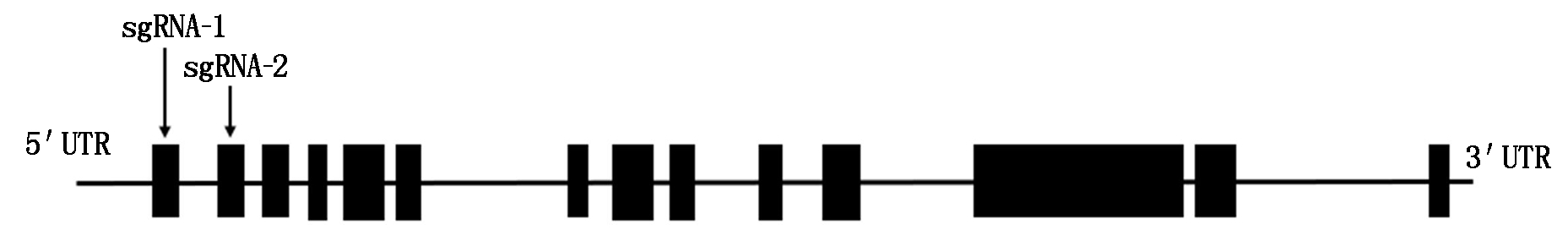
sgRNA-1. OsARF12第1个外显子靶位点;sgRNA-2. OsARF12第2个外显子靶位点。
sgRNA-1. The first exon target site of OsARF12;sgRNA-2. The second exon target site of OsARF12.
图1 OsARF12靶位点设计
Fig.1 Design of target sites of OsARF12
1.2.4 OsARF12表达量分析 将cDNA稀释10倍,使用天根生化科技有限公司定量检测试剂盒SYBR Green(FP209),Actin基因作为内参,采用Bio-Rad CFX96荧光定量PCR仪进行实时荧光定量PCR分析。相对表达量采用2-ΔΔCt法计算。
1.2.5 数据分析 采用Microsoft Excel 2016对试验数据进行整理和作图,利用SPSS 24进行差异显著性分析。
2 结果与分析
2.1 KO-ARF12-1/-2 T0突变体鉴定
将携带CRISPR/Cas9-ARF12-1和CRISPR/Cas9-ARF12-2质粒的农杆菌侵染水稻品种日本晴,得到KO-ARF12-1 T0再生苗5株,KO-ARF12-2 T0再生苗15株。利用CTAB法提取各单株叶片的DNA,并根据OsARF12的序列分别设计2个不同靶位点引物:Crispr ARF12-1_F、Crispr ARF12-1_R和Crispr ARF12-2_F、Crispr ARF12-2_R(表1),用对应的引物对转基因植株的DNA进行扩增(图2),并将扩增产物送往公司测序。通过对T0测序结果比对发现:KO-ARF12-1-1为1条链缺失1 nt、互补链缺失7 nt的双等位突变体;KO-ARF12-1-2和KO-ARF12-1-3为1条链缺失4 nt、互补链缺失7 nt的双等位突变体,KO-ARF12-1-4和KO-ARF12-1-5 2条链均未突变(表2)。
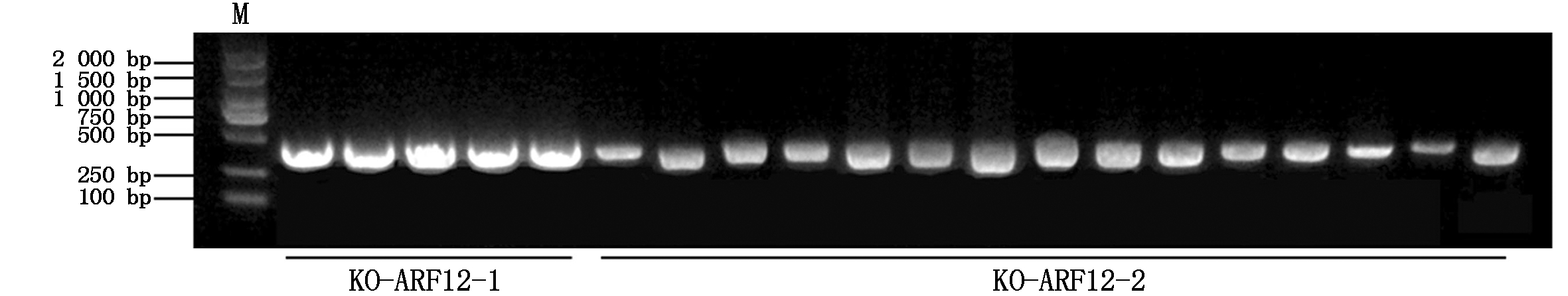
M.2 kb DNA 标准分子质量。图3-4同。
M.2 kb DNA Marker. The same as Fig.3-4.
图2 OsARF12 T0转基因植株鉴定
Fig. 2 Identification of OsARF12 T0 transgenic plants
表2 KO-ARF12-1 T0转基因植株突变类型鉴定
Tab.2 Identification of mutation types of KO-ARF12-1 T0 transgenic plants
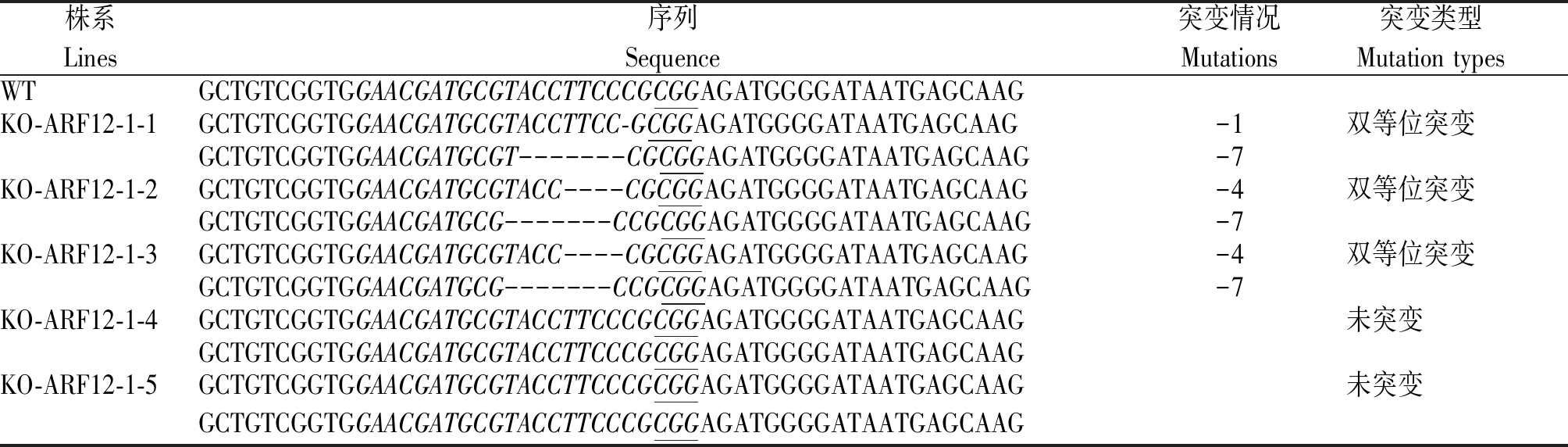
株系Lines序列Sequence突变情况Mutations突变类型Mutation typesWTGCTGTCGGTGGAACGATGCGTACCTTCCCGCGGAGATGGGGATAATGAGCAAGKO-ARF12-1-1GCTGTCGGTGGAACGATGCGTACCTTCC-GCGGAGATGGGGATAATGAGCAAG-1双等位突变GCTGTCGGTGGAACGATGCGT-------CGCGGAGATGGGGATAATGAGCAAG-7KO-ARF12-1-2GCTGTCGGTGGAACGATGCGTACC----CGCGGAGATGGGGATAATGAGCAAG-4双等位突变GCTGTCGGTGGAACGATGCG-------CCGCGGAGATGGGGATAATGAGCAAG-7KO-ARF12-1-3GCTGTCGGTGGAACGATGCGTACC----CGCGGAGATGGGGATAATGAGCAAG-4双等位突变GCTGTCGGTGGAACGATGCG-------CCGCGGAGATGGGGATAATGAGCAAG-7KO-ARF12-1-4GCTGTCGGTGGAACGATGCGTACCTTCCCGCGGAGATGGGGATAATGAGCAAG未突变GCTGTCGGTGGAACGATGCGTACCTTCCCGCGGAGATGGGGATAATGAGCAAGKO-ARF12-1-5GCTGTCGGTGGAACGATGCGTACCTTCCCGCGGAGATGGGGATAATGAGCAAG未突变GCTGTCGGTGGAACGATGCGTACCTTCCCGCGGAGATGGGGATAATGAGCAAG
注:斜体碱基. OsARF12靶序列;下划线碱基. PAM序列;短线. 碱基缺失。表3-5同。
Note:Italicized bases are the target sites of OsARF12;Underline bases are the PAM sequence;Short lines are the base deletion;The same as Tab. 3-5.
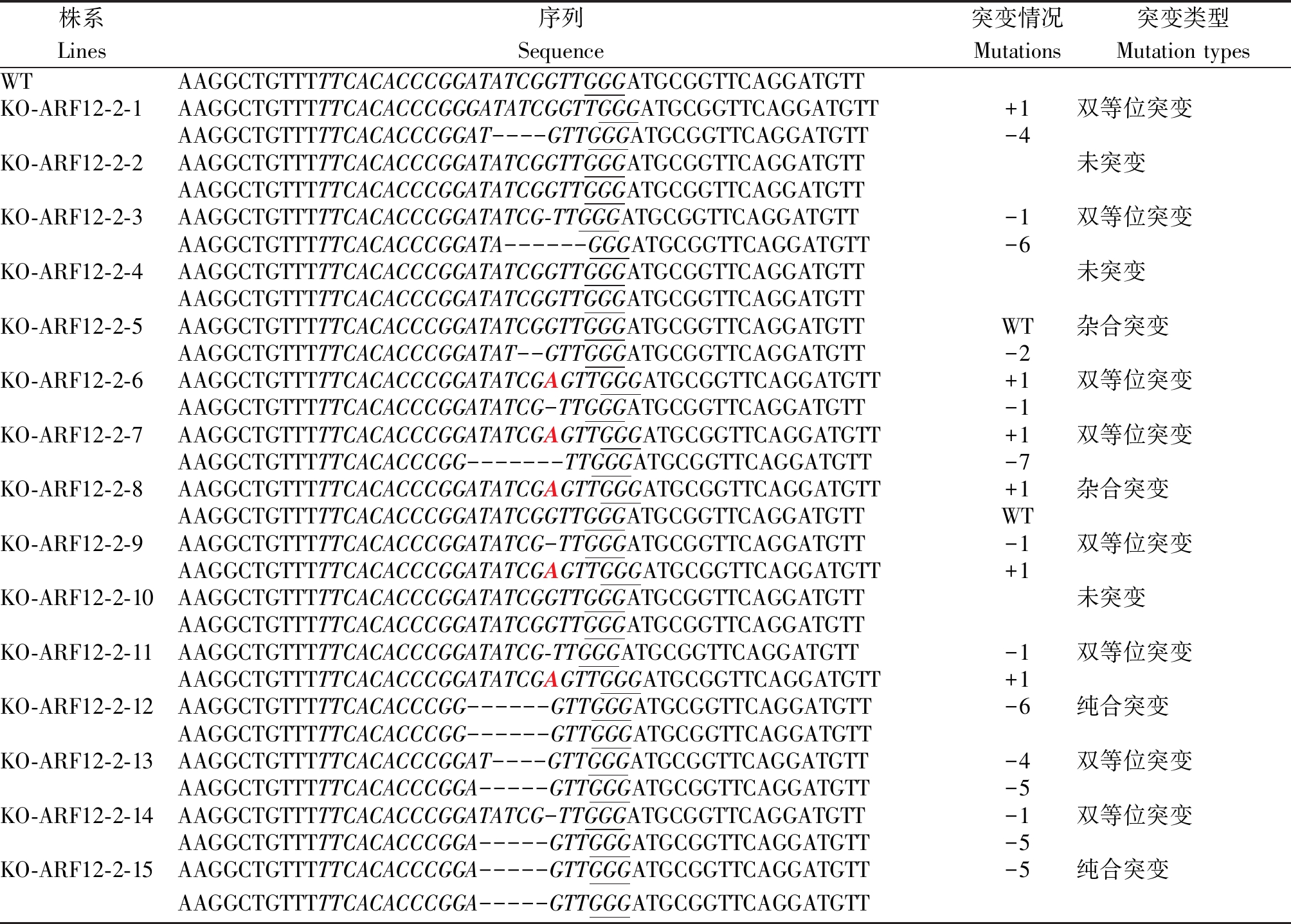
KO-ARF12-2各转基因植株共有11种突变类型:KO-ARF12-2-12和KO-ARF12-2-15为2条链分别缺失了6 nt与5 nt的纯合突变体;KO-ARF12-2-5为1条链缺失2 nt,KO-ARF12-2-8为1条链插入1 nt,互补链均没有突变的杂合突变体;而其余8株均为双等位突变:KO-ARF12-2-1为1条链插入1 nt,互补链缺失4 nt;KO-ARF12-2-3为1条链缺失1 nt,互补链缺失6 nt;KO-ARF12-2-6、KO-ARF12-2-9和KO-ARF12-2-11为1条链插入1 nt,互补链缺失1 nt;KO-ARF12-2-7为1条链插入1 nt,互补链缺失7 nt;KO-ARF12-2-13为1条链缺失4 nt,互补链缺失5 nt;KO-ARF12-2-14为1条链缺失1 nt,互补链缺失5 nt;KO-ARF12-2-2、KO-ARF12-2-4和KO-ARF12-2-10 2条链均未突变(表3)。
表3 KO-ARF12-2 T0转基因植株突变类型鉴定
Tab.3 Identification of mutation types of KO-ARF12-2 T0 transgenic plants
株系Lines序列Sequence突变情况Mutations突变类型Mutation typesWTAAGGCTGTTTTTCACACCCGGATATCGGTTGGGATGCGGTTCAGGATGTTKO-ARF12-2-1AAGGCTGTTTTTCACACCCGGGATATCGGTTGGGATGCGGTTCAGGATGTT+1双等位突变AAGGCTGTTTTTCACACCCGGAT----GTTGGGATGCGGTTCAGGATGTT-4KO-ARF12-2-2AAGGCTGTTTTTCACACCCGGATATCGGTTGGGATGCGGTTCAGGATGTT未突变AAGGCTGTTTTTCACACCCGGATATCGGTTGGGATGCGGTTCAGGATGTTKO-ARF12-2-3AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1双等位突变AAGGCTGTTTTTCACACCCGGATA------GGGATGCGGTTCAGGATGTT-6KO-ARF12-2-4AAGGCTGTTTTTCACACCCGGATATCGGTTGGGATGCGGTTCAGGATGTT未突变AAGGCTGTTTTTCACACCCGGATATCGGTTGGGATGCGGTTCAGGATGTTKO-ARF12-2-5AAGGCTGTTTTTCACACCCGGATATCGGTTGGGATGCGGTTCAGGATGTTWT杂合突变AAGGCTGTTTTTCACACCCGGATAT--GTTGGGATGCGGTTCAGGATGTT-2KO-ARF12-2-6AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1双等位突变AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1KO-ARF12-2-7AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1双等位突变AAGGCTGTTTTTCACACCCGG-------TTGGGATGCGGTTCAGGATGTT-7KO-ARF12-2-8AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1杂合突变AAGGCTGTTTTTCACACCCGGATATCGGTTGGGATGCGGTTCAGGATGTTWTKO-ARF12-2-9AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1双等位突变AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1KO-ARF12-2-10AAGGCTGTTTTTCACACCCGGATATCGGTTGGGATGCGGTTCAGGATGTT未突变AAGGCTGTTTTTCACACCCGGATATCGGTTGGGATGCGGTTCAGGATGTTKO-ARF12-2-11AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1双等位突变AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1KO-ARF12-2-12AAGGCTGTTTTTCACACCCGG------GTTGGGATGCGGTTCAGGATGTT-6纯合突变AAGGCTGTTTTTCACACCCGG------GTTGGGATGCGGTTCAGGATGTTKO-ARF12-2-13AAGGCTGTTTTTCACACCCGGAT----GTTGGGATGCGGTTCAGGATGTT-4双等位突变AAGGCTGTTTTTCACACCCGGA-----GTTGGGATGCGGTTCAGGATGTT-5KO-ARF12-2-14AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1双等位突变AAGGCTGTTTTTCACACCCGGA-----GTTGGGATGCGGTTCAGGATGTT-5KO-ARF12-2-15AAGGCTGTTTTTCACACCCGGA-----GTTGGGATGCGGTTCAGGATGTT-5纯合突变AAGGCTGTTTTTCACACCCGGA-----GTTGGGATGCGGTTCAGGATGTT
注:红色加粗倾斜碱基为碱基插入。表5同。
Note: Red bold tilt bases are bases insertion. The same as Tab. 5.
2.2 KO-ARF12-1/-2 T1突变体鉴定
采用CTAB法提取OsARF12在T0发生突变的转基因株系叶片DNA,并用Crispr ARF12-1_F、Crispr ARF12-1_R和Crispr ARF12-2_F、Crispr ARF12-2_R对相应的转基因株系DNA进行扩增(图3,4),将产物送往公司测序。测序结果进行比对发现,KO-ARF12-1各突变转基因株系T1 20个单株中,有3种纯合突变、2种双等位突变,共5种突变类型,分别为:1条链缺失1 nt、互补链缺失7nt的双等位突变体有4株;缺失1 nt的纯合突变体有1株;缺失7 nt的纯合突变体有3株;缺失4 nt的纯合突变体有7株;1条链缺失4 nt、互补链缺失7 nt的双等位突变体有5株(表4)。
图3 KO-ARF12-1 T1转基因植株鉴定
Fig.3 Identification of KO-ARF12-1 T1 transgenic plants
表4 KO-ARF12-1 T1转基因植株突变类型鉴定
Tab.4 Analysis and identification of mutation types of KO-ARF12-1 T1 transgenic plants

株系Lines序列Sequence突变情况Mutations突变类型Mutation typesWTGCTGTCGGTGGAACGATGCGTACCTTCCCGCGGAGATGGGGATAATGAGCAAGKO-ARF12-1-1-1GCTGTCGGTGGAACGATGCGTACCTTCC-GCGGAGATGGGGATAATGAGCAAG-1双等位突变GCTGTCGGTGGAACGATGCGT-------CGCGGAGATGGGGATAATGAGCAAG-7KO-ARF12-1-1-3GCTGTCGGTGGAACGCTGCGTACCTTCC-GCGGAGATGGGGATAATGAGCAAG-1纯合突变GCTGTCGGTGGAACGCTGCGTACCTTCC-GCGGAGATGGGGATAATGAGCAAGKO-ARF12-1-1-4GCTGTCGGTGGAACGATGCGT-------CGCGGAGATGGGGATAATGAGCAAG-7纯合突变GCTGTCGGTGGAACGATGCGT-------CGCGGAGATGGGGATAATGAGCAAGKO-ARF12-1-2-1GCTGTCGGTGGAACGATGCGTACC----CGCGGAGATGGGGATAATGAGCAAG-4纯合突变GCTGTCGGTGGAACGATGCGTACC----CGCGGAGATGGGGATAATGAGCAAGKO-ARF12-1-2-4GCTGTCGGTGGAACGATGCGTACC----CGCGGAGATGGGGATAATGAGCAAG-4双等位突变GCTGTCGGTGGAACGATGCG-------CCGCGGAGATGGGGATAATGAGCAAG-7KO-ARF12-1-2-6GCTGTCGGTGGAACGATGCG-------CCGCGGAGATGGGGATAATGAGCAAG-7纯合突变GCTGTCGGTGGAACGATGCG-------CCGCGGAGATGGGGATAATGAGCAAGKO-ARF12-1-3-1GCTGTCGGTGGAACGATGCGTACC----CGCGGAGATGGGGATAATGAGCAAG-4纯合突变GCTGTCGGTGGAACGATGCGTACC----CGCGGAGATGGGGATAATGAGCAAGKO-ARF12-1-3-6GCTGTCGGTGGAACGATGCGTACC----CGCGGAGATGGGGATAATGAGCAAG-4双等位突变GCTGTCGGTGGAACGATGCG-------CCGCGGAGATGGGGATAATGAGCAAG-7
图4 KO-ARF12-2 T1转基因植株鉴定
Fig.4 Identification of KO-ARF12-2 T1 transgenic plants
KO-ARF12-2各突变转基因株系T1 60个单株中,有6种纯合突变、1种杂合突变和4种双等位突变,共11种突变类型,分别为:缺失1,6,2,7,4 nt和插入1 nt的纯合突变体,分别为12,1,6,3,2,10株;1条链插入1 nt、互补链未突变的杂合突变体3株;1条链插入1 nt、互补链缺失4 nt,1条链插入1 nt、互补链缺失1 nt,1条链缺失4 nt、互补链缺失5 nt和1条链缺失1 nt、互补链缺失5 nt的双等位突变体分别为6,9,4,4株(表5)。
2.3 KO-ARF12-1/-2 T2突变体表达量分析
以T2 2种不同靶位点的6种不同基因型纯合突变体KO-ARF12-1-1-3(缺失1 nt)、KO-ARF12-1-2-1(缺失4 nt)、KO-ARF12-1-2-6(缺失7 nt)及KO-ARF12-2-5-2(缺失2 nt)、KO-ARF12-2-11-4(插入1 nt)、KO-ARF12-2-15-5(缺失5 nt)(图5-A)为研究材料,选取大田正常生长条件下叶片样品提取总RNA,反转录并定量分析发现,除KO-ARF12-2-11-4,其他各突变体株系中OsARF12表达量与野生型相比均显著下降(P<0.05)(图5-B)。
表5 KO-ARF12-2 T1转基因植株突变类型鉴定
Tab.5 Analysis and identification of mutation types of KO-ARF12-2 T1 transgenic plants

株系Lines序列Sequence突变情况Mutations突变类型Mutation typesWTAAGGCTGTTTTTCACACCCGGATATCGGTTGGGATGCGGTTCAGGATGTTKO-ARF12-2-1-1AAGGCTGTTTTTCACACCCGGGATATCGGTTGGGATGCGGTTCAGGATGTT+1双等位突变AAGGCTGTTTTTCACACCCGGAT----GTTGGGATGCGGTTCAGGATGTT-4KO-ARF12-2-3-2AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1纯合突变AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTTKO-ARF12-2-3-5AAGGCTGTTTTTCACACCCGGATA------GGGATGCGGTTCAGGATGTT-6纯合突变AAGGCTGTTTTTCACACCCGGATA------GGGATGCGGTTCAGGATGTTKO-ARF12-2-5-2AAGGCTGTTTTTCACACCCGGATAT--GTTGGGATGCGGTTCAGGATGTT-2纯合突变AAGGCTGTTTTTCACACCCGGATAT--GTTGGGATGCGGTTCAGGATGTTKO-ARF12-2-6-1AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1双等位突变AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1KO-ARF12-2-7-4AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1纯合突变AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTTKO-ARF12-2-7-1AAGGCTGTTTTTCACACCCGG-------TTGGGATGCGGTTCAGGATGTT-7纯合突变AAGGCTGTTTTTCACACCCGG-------TTGGGATGCGGTTCAGGATGTTKO-ARF12-2-8-1AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1杂合突变AAGGCTGTTTTTCACACCCGGATATCGGTTGGGATGCGGTTCAGGATGTTWTKO-ARF12-2-8-2AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1纯合突变AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTTKO-ARF12-2-9-1AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1纯合突变AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT
表5(续)

株系Lines序列Sequence突变情况Mutations突变类型Mutation typesKO-ARF12-2-9-2AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1纯合突变AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTTKO-ARF12-2-11-1AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1纯合突变AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTTKO-ARF12-2-11-2AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1双等位突变AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1KO-ARF12-2-11-4AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTT+1纯合突变AAGGCTGTTTTTCACACCCGGATATCGAGTTGGGATGCGGTTCAGGATGTTKO-ARF12-2-13-1AAGGCTGTTTTTCACACCCGGATAT----TGGGATGCGGTTCAGGATGTT-4纯合突变AAGGCTGTTTTTCACACCCGGATAT----TGGGATGCGGTTCAGGATGTTKO-ARF12-2-13-5AAGGCTGTTTTTCACACCCGGAT----GTTGGGATGCGGTTCAGGATGTT-4双等位突变AAGGCTGTTTTTCACACCCGGA-----GTTGGGATGCGGTTCAGGATGTT-5KO-ARF12-2-14-1AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1纯合突变AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTTKO-ARF12-2-14-5AAGGCTGTTTTTCACACCCGGATATCG-TTGGGATGCGGTTCAGGATGTT-1双等位突变AAGGCTGTTTTTCACACCCGGA-----GTTGGGATGCGGTTCAGGATGTT-5
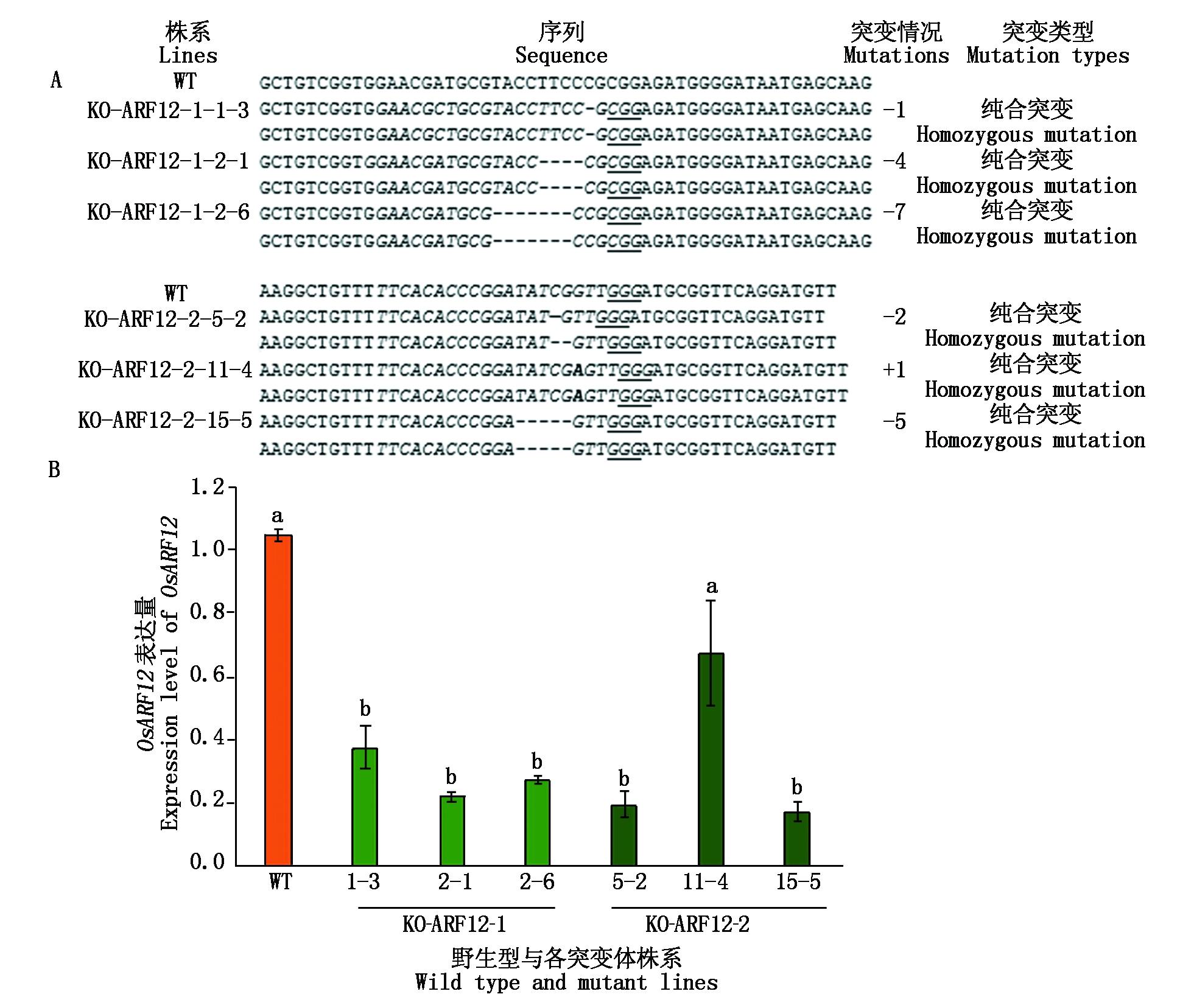
A. 突变体突变类型;B. 野生型和突变体中OsARF12表达量;不同字母表示不同株系间达到5%显著差异水平。图6同。
A. Mutation types of mutants;B. Expression levels of OsARF12 in wild type and mutants;
Different letters indicated that significant difference among different lines(P<0.05).The same as Fig. 6.
图5 OsARF12突变体突变类型及表达量分析
Fig.5 Analysis of mutation types and OsARF12 expression levels of mutants
2.4 OsARF12对水稻株高的影响
为研究OsARF12对水稻农艺性状的影响,于灌浆期将突变体株系和野生型株系进行比较,结果表明,KO-ARF12-1/-2的株高均显著降低(图6-A、C),各突变体株系株高与野生型相比分别减少了16.59%,16.63%,16.06%和14.94%,13.15%,18.39%(图6-C)。进一步对水稻各茎节间长度进行统计分析发现,与野生型相比,KO-ARF12-1/-2显著降低了第3和第4茎节间长度,其中第4茎节间降低幅度最大(图6-B、D),降低的幅度为23.46%(P<0.05),其次为第3茎节间,降低的幅度为21.91%(P<0.05),而第1和第2茎节间长度与对照无显著差异(P>0.05)。
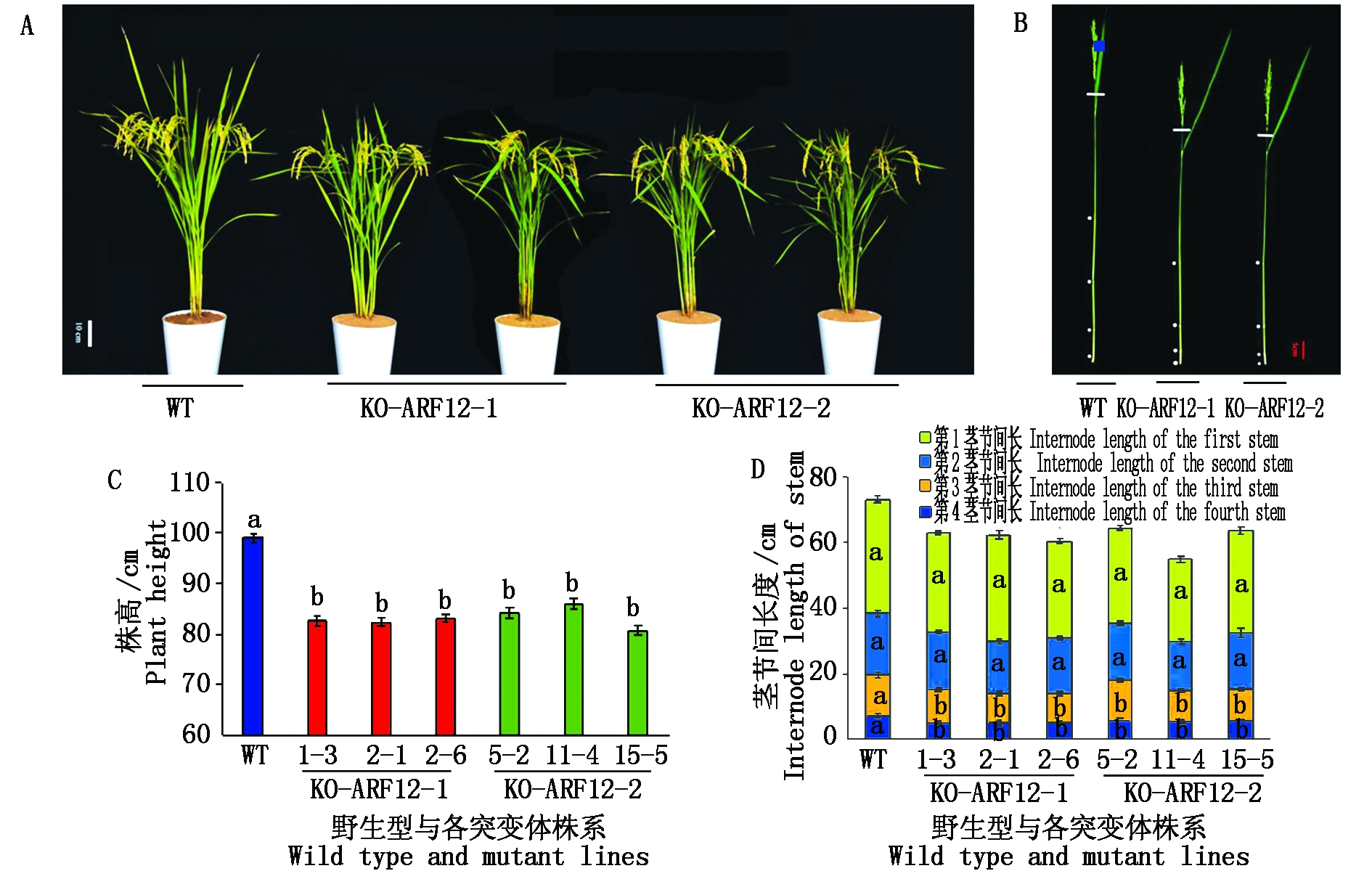
A-B. OsARF12突变体和野生型表型对比;
C-D. 突变体和野生型株高、茎节间长度分析;D中不同茎节间长度的方差分析分开进行。
A-B. Phenotype comparison of OsARF12 mutants and wild type;C-D. Analysis of plant height and internode length of mutants and wild type;The analysis of variance of different internode length in D was carried out separately.
图6 OsARF12对水稻株高的影响
Fig.6 Effects of OsARF12 on plant height
3 结论与讨论
现代农业发展中,对突变体进行基因功能研究已经成为一种重要的研究手段。2013年CRISPR/Cas9技术成功应用于定点突变水稻基因[23],2014年CRISPR/Cas9技术被证实可高效编辑水稻特异基因,并且基因突变能够稳定遗传[22],极大地促进了水稻突变体库的建立。本研究利用CRISPR/Cas9技术分别对OsARF12的第1、第2个外显子靶序列进行定点编辑,获得了OsARF12多种类型的突变体。其中,KO-ARF12-1 T0 3种突变类型,T1 5种突变类型;KO-ARF12-2 T0 11种突变类型,T1 11种突变类型,为研究OsARF12对水稻农艺性状影响奠定基础。
通过对KO-ARF12-1/-2 2种突变体材料中OsARF12的表达量进行鉴定发现,CRISPR/Cas9基因编辑技术显著降低了OsARF12的表达量。且与对照相比,KO-ARF12-1/-2株高显著降低(P<0.05),表明OsARF12在一定程度上正向调节水稻的株高性状。已有研究借助T-DNA插入或者是Tos17插入的arf12突变体研究发现,OsARF12可能介导根形态和磷诱导的生长素信号反应;Qi等[24]发现,OsARF12改变铁调节蛋白(OsMIR)和铁调节转运蛋白1(OsIRT1)的丰度,导致铁含量的改变。本研究通过CRISPR/Cas9基因编辑技术创制了OsARF12突变体,研究了其在调节水稻株高方面的重要作用,丰富了水稻生长素响应因子OsARF12的生物学功能。
水稻的株高受众多激素以及它们之间的相互作用调控,OsMADS57功能的缺失降低了GA的活性,导致株高降低[25];OsMED14_1与转录因子YABBY5、TDR和MADS29互作,参与调节生长素的动态平衡,进而调控水稻株高[26];miR1848靶向OsCYP51G3调节水稻中植物甾醇和BR的生物合成,影响水稻株高[27]。本研究通过CRISPR/Cas9技术对生长素响应因子OsARF12进行编辑,可能通过影响其生长素调控路径,进而导致株高降低。水稻株高由各茎节间长度组成,水稻抗倒伏能力与近基部茎节间长度呈负相关关系[28]。本研究表明,各突变类型的突变体与野生型相比,第1和第2茎节间长度无显著差异,第3和第4茎节间长度显著(P<0.05)降低,且第4茎节间降幅最大,表明突变体植株越靠近基部,其茎节间长度缩短幅度越大,这对提高水稻的抗倒伏能力有重要作用,但KO-ARF12突变体中不同节间长度改变的内在机制需要进一步研究。
[1] Ljung K. Auxin metabolism and homeostasis during plant development[J]. Development ,2013,140(5):943-950. doi:10.1242/dev.086363.
[2] Bohn-Courseau I. Auxin:A major regulator of organogenesis[J]. Comptes Rendus Biologies,2010,333(4):290-296. doi:10.1016/j.crvi.2010.01.004.
[3] 蒋素梅,陶均,李玲. 早期生长素响应蛋白在生长素信号转导中的作用[J].植物生理学通讯,2005,41(1):125-130.
Jiang S M,Tao J,Li L. The roles of early auxin response proteins in auxin signal transduction[J]. Plant Physiology Communications,2005,41(1):125-130.
[4] Ulmasov T,Hagen G,Guilfoyle T J. ARF1,a transcription factor that binds to auxin response elements[J]. Science,1997,276(5320):1865-1868. doi:10.1126/science.276.5320.1865.
[5] Okushima Y,Overvoorde P J,Arima K,Alonso J M,Chan A,Chang C,Ecker J R,Hughes B,Lui A,Nguyen D,Onodera C,Quach H,Smith A,Yu G X,Theologis A. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana:Unique and overlapping functions of ARF7 and ARF19[J]. The Plant Cell,2005,17(2):444-463. doi:10.1105/tpc.104.028316.
[6] Ellis C M,Nagpal P,Young J C,Hagen G,Guilfoyle T J,Reed J W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana[J]. Development,2005,132(20):4563-4574. doi:10.1242/dev.02012.
[7] Pekker I,Alvarez J P,Eshed Y. Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity[J]. The Plant Cell,2005,17(11):2899-2910. doi:10.1105/tpc.105.034876.
[8] Cheng Z J,Wang L,Sun W, Zhang Y,Zhou C,Su Y H,Li W,Sun T T,Zhao X Y,Li X G,Cheng Y F,Zhao Y D,Xie Q,Zhang X S. Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3[J]. Plant Physiology,2013,161(1):240-251. doi:10.1104/pp.112.203166.
[9] Hardtke C S,Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development [J]. The EMBO Journal,1998,17(5):1405-1411. doi:10.1093/emboj/17.5.1405.
[10] Nagpal P,Ellis C M,Weber H,Ploense S E,Barkawi L S,Guilfoyle T J,Hagen G,Alonso J M,Cohen J D,Farmer E E,Ecker J R,Reed J W. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation[J].Development,2005,132(18):4107-4118. doi:10.1242/dev.01955.
[11] Goetz M,Vivian-Smith A,Johnson S D,Koltunow A M. AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis[J]. The Plant Cell,2006,18(8):1873-1886. doi:10.1105/tpc.105.037192.
[12] Okushima Y,Fukaki H,Onoda M,Theologis A,Tasaka M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis[J]. The Plant Cell,2007,19(1):118-130. doi:10.1105/tpc.106.047761.
[13] Wang J W,Wang L J,Mao Y B,Cai W J,Xue H W,Chen X Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis[J]. The Plant Cell,2005,17(8):2204-2216. doi:10.1105/tpc.105.033076.
[14] Wang D K,Pei K M,Fu Y P,Sun Z X,Li S J,Liu H Q,Tang K,Han B,Tao Y Z. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa)[J]. Gene,2007,394(1/2):13-24. doi:10.1016/j.gene.2007.01.006.
[15] Waller F,Furuya M,Nick P. OsARF1,an auxin response factor from rice,is auxin-regulated and classifies as a primary auxin responsive gene[J]. Plant Molecular Biology,2002,50(3):415-425. doi:10.1023/A:1019818110761.
[16] Attia K A,Abdelkhalik A F,Ammar M H,Wei C,Yang J S,Lightfoot D A,El-Sayed W M,El-Shemy-H A. Antisense phenotypes reveal a functional expression of OsARF1,an auxin response factor,in transgenic rice[J]. Current Issues in Molecular Biology,2009,11(S1):29-34. doi:10.21775/9781912530069.04.
[17] Hu Z J,Lu S J,Wang M J,He H H,Sun L,Wang H R,Liu X H,Jiang L,Sun J L,Xin X Y,Kong W,Chu C C,Xue H W,Yang J S,Luo X J,Liu J X. A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice [J]. Molecular Plant,2018,11(5):736-749. doi:10.1016/j.molp.2018.03.005.
[18] Shen C J,Yue R Q,Yang Y J,Zhang L,Sun T,Tie S G,Wang H Z. OsARF16 is involved in cytokinin-mediated inhibition of phosphate transport and phosphate signaling in rice (Oryza sativa L.)[J]. PLoS One,2014,9(11):e112906. doi:10.1371/journal.pone.0112906.
[19] Chen S H,Zhou L J,Xu P,Xue H W. SPOC domain-containing protein Leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling[J]. PLoS Genetics,2018,14(11):e1007829. doi:10.1371/journal.pgen.1007829.
[20] Zhang S N,Wang S K,Xu Y X,Yu C L,Shen C J,Qian Q,Geisler M,Jiang D A,Qi Y H. The auxin response factor,OsARF19,controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1[J]. Plant Cell & Environment,2015,38(4):638-654. doi:10.1111/pce.12397.
[21] Ran F A, Hsu P D, Wright J, Agarwala V, Scott D A, Zhang F. Genome engineering using the CRISPR-Cas9 system[J]. Nature Protocols,2013,8(2):2281-2308. doi:10.1038/nprot.2013.143.
[22] Zhang H,Zhang J S,Wei P L,Zhang B T,Gou F,Feng Z Y,Mao Y F,Yang L,Zhang H,Xu N F,Zhu J K. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation[J]. Plant Biotechnology Journal,2014,12(6):797-807. doi:10.1111/pbi.12200.
[23] Feng Z Y,Zhang B T,Ding W N,Liu X D,Yang D L,Wei P L,Cao F Q,Zhu S H,Zhang F,Mao Y F,Zhu J K. Efficient genome editing in plants using a CRISPR/Cas system[J].Cell Research,2013,23(10):1229-1232. doi:10.1038/cr.2013.114.
[24] Qi Y H,Wang S K,Shen C J,Zhang S N,Chen Y,Xu Y X,Liu Y,Wu Y R,Jiang D A. OsARF12, a transcription activator on auxin response gene,regulates root elongation and affects iron accumulation in rice (Oryza sativa)[J].The New Phytologist,2012,193(1):109-120. doi:10.1111/j.1469-8137.2011.03910.x.
[25] Chu Y L,Xu N,Wu Q,Yu B,Li X X,Chen R R,Huang J L. Rice transcription factor OsMADS57 regulates plant height by modulating gibberellin catabolism[J].Rice,2019,12(1):1-14. doi:10.1186/s12284-019-0298-6.
[26] Malik N,Ranjan R,Parida S K,Agarwal P,Tyagi A K. Mediator subunit OsMED14_1 plays an important role in rice development [J].The Plant Journal,2020,101(6):1411-1429. doi:10.1111/tpj.14605.
[27] Xia K F,Ou X J,Tang H D,Wang R,Wu P,Jia Y X,Wei X Y,Xu X L,Kang S H,Kim S K,Zhang M Y. Rice microRNA osa-miR1848 targets the obtusifoliol 14α-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress[J]. The New Phytologist,2015,208(3):790-802. doi:10.1111/nph.13513.
[28] Kashiwagi T,Ishimaru K. Identification and functional analysis of a locus for improvement of lodging resistance in rice[J].Plant Physiology,2004,134(2):676-683. doi:10.1104/pp.103.029355.