栽培地土壤盐分过高,土质盐碱化成为植株生长和发育进程中的主要环境限制因子之一[1-3]。高盐胁迫下植株生长环境改变,过多钠离子(Na+)在植株体内累积,导致质膜完整性和通透性丧失[4],正常生理活动被抑制,进而造成植物早衰加速,品质和产量下降[5-6]。研究表明,Na+/H+逆向转运蛋白(Vacuolar Na+/H+ exchange or antiporter,NHX)在植株体内不仅参与细胞扩增[7]、pH值调节[8]、保护K+稳态性[9-10],还可参与到抗盐胁迫[11]和Na+长距离运输[12]中可将植株细胞内多余吸收的Na+向外排出或在液泡中对Na+进行区域化来控制Na+累积对膜系统的伤害,是植株体内抵御盐胁迫、维持离子稳定、调节细胞渗透重要的参与者[13-14]。
在甜菜(Beta vulgaris)[15]的下部根系中首次检测到液泡膜型Na+/H+逆向转运蛋白(NHX)活性的发生,随后在拟南芥(Arabidopsis thaliana)[16]中发现植物中最早的液泡膜型Na+/H+逆向转运蛋白基因AtNHX1的存在。目前已在玉米(Zea mays)[17]、盐地碱蓬(Suaeda salsa)[18]、菊苣(Cichorium intybus L.)[19]、豇豆(Vigna unguiculata L.)[20]等多种植物中克隆到液泡膜型Na+/H+逆向转运蛋白基因NHX的同源基因,说明NHX1在高等植物生物进程中普遍存在[4,21],且通过异源表达酵母互补试验证实,NHX1基因不仅对酵母中Na+转运起到直接调节作用,而且其耐盐性也有所提高[22]。Fukuda等[23]研究发现,在盐胁迫下,基因OsNHX1的产物可作为Na+/H+交换子,并在水稻的耐盐性中起重要作用。郭会敏等[24]在200 mmol/L NaCl中进行NnNHX1 基因转化烟草耐盐性研究发现,转基因植株较对照有着良好的生长,且体内叶绿素含量显著高于对照,膜完整性更好,耐盐性提高。Zhang 等[25]在油菜研究中发现,过表达AtNHX1(来自拟南芥的液泡Na + /H + antiport)的转基因甘蓝型油菜植物能够在200 mmol/L NaCl处理下进行正常的生理进程,其耐盐性增加,且在油菜种子采收后其产量和菜籽油的品质在盐胁迫下无影响。He等[26]研究棉花表明,在200 mmol/L NaCl胁迫下,过表达AtNHX1基因的棉花植株较野生型相比有着更好光合作用性能和更高的氮同化率,且其产量和棉花纤维品质提高,转基因植株棉花的耐盐胁迫能力提高。Bao等[27]研究百脉根时发现,过表达ZxNHX转基因百脉根在200 mmol/L NaCl处理下较野生型表现出更好的保水能力和较高的净光合作用率,其对盐分的耐受性增强,植株生物量在后期生长中也有所增加。Su等[28]研究认为,高盐胁迫下0.2 mg/L外源油菜素内酯(BL)的施用增加了苹果幼苗体内反向转运蛋白基因(MhNHXs)的表达水平,提高了植株耐盐能力,有效减少了盐胁迫下芽和根中Na +的积累,保持了苹果幼苗体内渗透平衡。拟南芥AtNHX1在番茄[29]和菜籽[30]中超表达增加它们在高盐环境中的抗性。Long等[31]发现,GhNHX1的沉默导致棉花幼苗对高盐浓度的敏感性增强,这表明GhNHX1正调控棉花对盐胁迫的耐受性。Mushke等[32]研究向日葵(Helianthus annuus L.)耐盐性时发现,小麦TaNHX2基因过表达使得转基因向日葵植物显示出更好的抗细胞损伤保护能力,稳定的相对水含量、叶绿素含量、脯氨酸积累增加和活性氧物种(ROS)清除能力提高,其对盐分的抗性增强。Wu等[33]发现,AtNHX5在大豆中的超表达使得其在300 mmol/L NaCl胁迫下有着较强的耐受性。
目前,NHX耐盐机制虽然在多种植物当中都有研究,然而在辣椒(Capsicum annuum L.)这种日常调味蔬菜中有关液泡膜Na+/H+逆向转运蛋白NHX耐盐性的作用机制研究尚不清楚,通过了解辣椒中NHX基因家族生物信息学功能,探究其在非生物胁迫下基因的表达特性,进而为辣椒CaNHX基因的功能开发以及辣椒耐盐抗旱育种基因提供基础。
1 材料和方法
1.1 试验材料
以当地日光温室主栽辣椒品种陇椒5号为非生物胁迫处理材料。在陇椒5号辣椒幼苗真叶完全展开叶多于6片时进行[34]。
胁迫处理:渗透胁迫(PEG )利用5%浓度甘露醇,赤霉素 (GA3) 、茉莉酸甲酯 (MeJA) 和NaCl分别以浓度100,200,200 mmol/L喷施在辣椒幼苗真叶的正反面,对照以蒸馏水喷洒处理。辣椒幼苗植株培养在12 h光照(光照强度为200 μmol/(m2·s))和12 h黑暗的培养箱内;白天温度设置为28 ℃,晚上设置为26 ℃[31]。
1.2 试验方法
1.2.1 辣椒幼苗真叶总RNA的提取与cDNA的合成 总RNA的提取:按照PureLink RNA小量提取试剂盒说明书对幼苗真叶中RNA进行提取。
RNA逆转录为cDNA的步骤:利用mRNA模板按照InvitrogenTM SuperScriptTM试剂盒说明进行。
1.2.2 CaNHXs家族基因成员的筛索 利用拟南芥基因库(https://www.arabidopsis.org)和茄科植物基因库(https://solgenomics.net)搜索删选符合辣椒NHX相应序列的基因,确定其NHX基因家族的成员。
1.2.3 CaNHXs家族基因生物信息特性分析 利用ProtParam[34]对CaNHXs家族基因理化特质进行计算。NHX基因的信号肽、蛋白亲疏水性以及跨膜结构分别通过SignalP4.1 Server、ProtScale和TMHMM进行预测。CaNHXs家族蛋白的保守结构域和亚细胞定位分析分别利用MEME和WoLF PSORT[35]进行。通过FGENESH-C[36]对序列进行基因框架预测。CaNHXs家族蛋白二级结构对比分析通过Prabi。基因顺式作用元件分析利用PlantCARE软件进行。通过在线软件phyre(http://www.sbg.bio.ic.ac.uk/)对NHX基因的三级结构进行预测。MEGA 5.0[37]和ClustalX[38]软件对NHX基因进行系统发育树绘制。
1.2.4 CaNHXs家族基因的定量表达分析 提取辣椒真叶叶片RNA,利用试剂盒反转录出cDNA,通过软件Primer premier 5.0以辣椒Actin基因作内参设计引物,通过KAPA SYBR® FAST定量荧光仪,以不同非生物胁迫处理下辣椒真叶叶片cDNA为试材,对非生物胁迫处理CaNHXs基因家族进行定量荧光差异分析。差异表达量通过2-ΔΔCt法[39]计算得出。
定量荧光所用引物序列见表1。
表1 CaNHXs基因家族表达分析的实时荧光定量引物
Tab.1 qRT-PCR primers for expression on analysis of CaNHXs

基因 Gene 实时荧光定量上游引物(5′-3′)Forward primer for qRT-PCR实时荧光定量下游引物(5′-3′)Reverse primer for qRT-PCRCaNHX1AAGAAGCTGTATTTTGGCAGGTGTGCCAAGTGTAATGAGACACaNHX2GAGGTCATACTCAGTTACGTGGCGTCGGTTCAGAAGAGATCACaNHX3ACCTTTCGTATGTAATGGCAGC CAGTGTCGCAAAAGCATGTCaNHX4CGTGACTACCTTGGTATTGGA CATCAATCTTCTGCACCGCACaNHX5GGGCATTGATATCGCGATGGAA GTGCTTTTTGATGTTTTGCTGCaNHX6GCATAGAACTGCACGACAG CGCATAAGTAACTTGTCTGGTCaNHX7GAACGATGGGGCAGCTATTGCCGAAGCCAGAAAAGAGATGACTINAGGGATGGGTCAAAAGGATGCGAGACAACACCGCCTGAATAGC
1.2.5 分析与绘图 通过SPSS 22软件对测得数据进行整理分析,利用Excel 2010绘图,定量试验均进行3次重复。
2 结果与分析
2.1 CaNHXs基因家族信息及蛋白理化性质分析
辣椒7个NHX基因通过ProtParam进行定位分析表明(表2):7个辣椒NHX基因各自分布在5条染色体上。其基因大小之间差异明显,CaNHX5的基因全长最小(1 231 bp),CaNHX6的基因全长最长(11 897 bp)。CaNHXs的氨基酸数目维持在198~952,CaNHX5的氨基酸数目最小,为198 aa,CaNHX7的氨基酸数目最大,为952 aa。CaNHX5的相对分子质量最小,为21.868 57 ku,CaNHX7的相对分子质量最大,为105.113 89 ku。各基因等电点变化范围差异较大,分布在5.53~9.18。CaNHX4和CaNHX7的不稳定系数均大于40,说明这2个基因编码产物不稳定。
表2 辣椒NHX基因家族信息及理化性质
Tab.2 Physical and chemical property of CaNHX
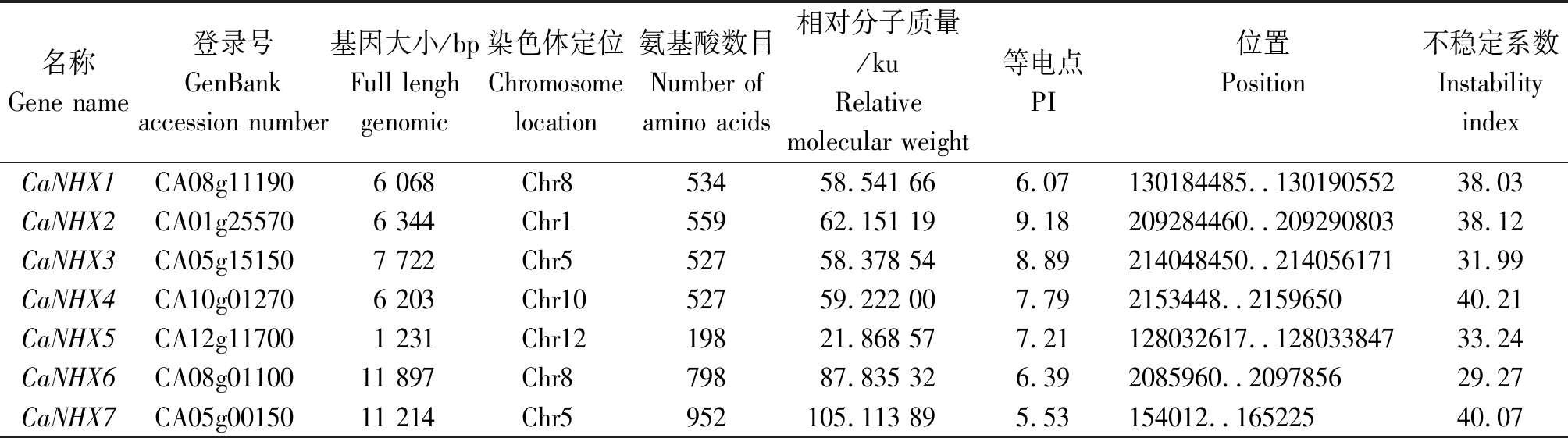
名称Gene name 登录号GenBank accession number 基因大小/bpFull lenghgenomic染色体定位Chromosome location氨基酸数目Number of amino acids相对分子质量/kuRelativemolecular weight等电点PI位置Position不稳定系数InstabilityindexCaNHX1CA08g111906 068Chr853458.541 666.07130184485..13019055238.03CaNHX2CA01g255706 344Chr155962.151 199.18209284460..20929080338.12CaNHX3CA05g151507 722Chr552758.378 548.89214048450..21405617131.99CaNHX4CA10g012706 203Chr1052759.222 007.792153448..215965040.21CaNHX5CA12g117001 231Chr1219821.868 577.21128032617..12803384733.24CaNHX6CA08g0110011 897Chr879887.835 326.392085960..209785629.27CaNHX7CA05g0015011 214Chr5952105.113 895.53154012..16522540.07
2.2 CaNHXs蛋白的亲水性/疏水性、跨膜结构与亚细胞定位
通过Signal P4.1 Server对辣椒CaNHXs基因家族成员所含有的信号肽预测分析发现,7个NHX蛋白中CaNHX1~CaNHX4、CaNHX6和CaNHX7均不具有信号肽,因此,这6个蛋白都不属于分泌蛋白类型,而CaNHX5则相反,在17~18号氨基酸有信号肽属于分泌蛋白类型。
利用ProtScal软件对CaNHXs基因家族成员进行亲水性/疏水性的预测,结果表明,7个NHX蛋白多肽链不同位置(CaNHX1为374和65位,CaNHX2为529和35位,CaNHX3为465和26位,CaNHX4为450和32位,CaNHX5为149和120位,CaNHX6为636和779位,CaNHX7为339和56位)出现亲水性(精氨酸(Arg))和疏水性(异亮氨酸(Ile))最强的氨基酸都相同且分值大小相近,7个NHX蛋白的平均亲疏水性值预测结果分别是,CaNHX1为0.572,CaNHX2为0.543,CaNHX3为0.512,CaNHX4为0.468,CaNHX5为0.708,CaNHX6为0.356,CaNHX7为0.259。说明NHX整条多肽链表现为疏水性,故推测CaNHXs蛋白为疏水性蛋白。
通过TMHMM对辣椒NHX蛋白进行跨膜结构预测分析得到CaNHXs基因家族成员有明显的跨膜结构,均属于跨膜蛋白。
对CaNHXs基因亚细胞定位预测表明(表3),CaNHXs基因家族成员在细胞膜(除CaNHX5)、内质网和液泡中均有不同程度的表达;在线粒体中表达的有CaNHX1、 CaNHX2 、CaNHX4;在细胞核中只有CaNHX3有表达。
表3 辣椒NHX基因的亚细胞定位
Tab.3 Subcellular location prediction of NHX genes in pepper
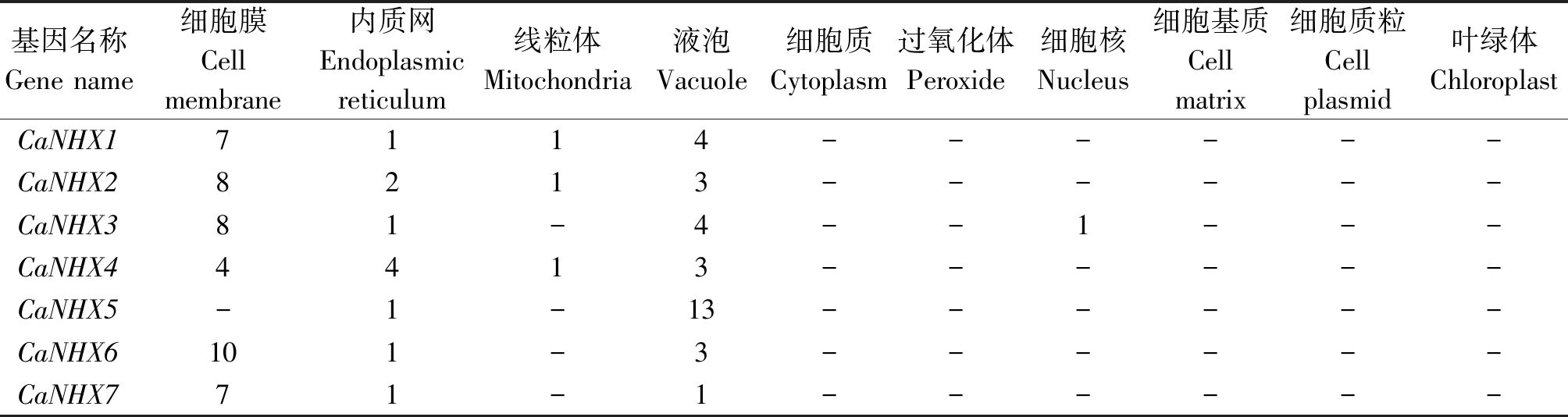
基因名称Gene name细胞膜Cellmembrane内质网Endoplasmic reticulum线粒体Mitochondria液泡Vacuole细胞质Cytoplasm过氧化体Peroxide细胞核Nucleus细胞基质Cell matrix细胞质粒Cell plasmid叶绿体ChloroplastCaNHX17114------CaNHX28213------CaNHX381-4--1---CaNHX44413------CaNHX5-1-13------CaNHX6101-3------CaNHX771-1------
注:括号内的数值代表预测得分。
Note:The numbers in parenthesis mean the prediction scores of protein locatio.
2.3 CaNHXs基因家族保守结构域分析
通过MEME软件对辣椒NHX基因成员结构域预测,共得到10个结构域,都包含有Na+/H+exchange保守结构域,其中CaNHX5 中只含有Motif5,CaNHX6中只含有Motif9,CaNHX7中只含有Motif6和Motif9存在,其他4个基因的保守结构域均相似,具有9个Motif的存在,N端主要是Motif7,该氨基酸保守域结构含有CXC24XC的锌指结构,C端主要是Motif6,该氨基酸保守域结构含有Trp(W)-24和TrKA-N,在序列中高度保守(图1)。对每个结构域进行分析发现(图2),Motif1、Motif2、Motif3、Motif4、Motif7、Motif9均含有50个保守氨基酸,Motif5含有49个保守氨基酸,Motif6含有41个保守氨基酸,Motif8含有40个保守氨基酸,Motif10含有的保守氨基酸较少有23个。
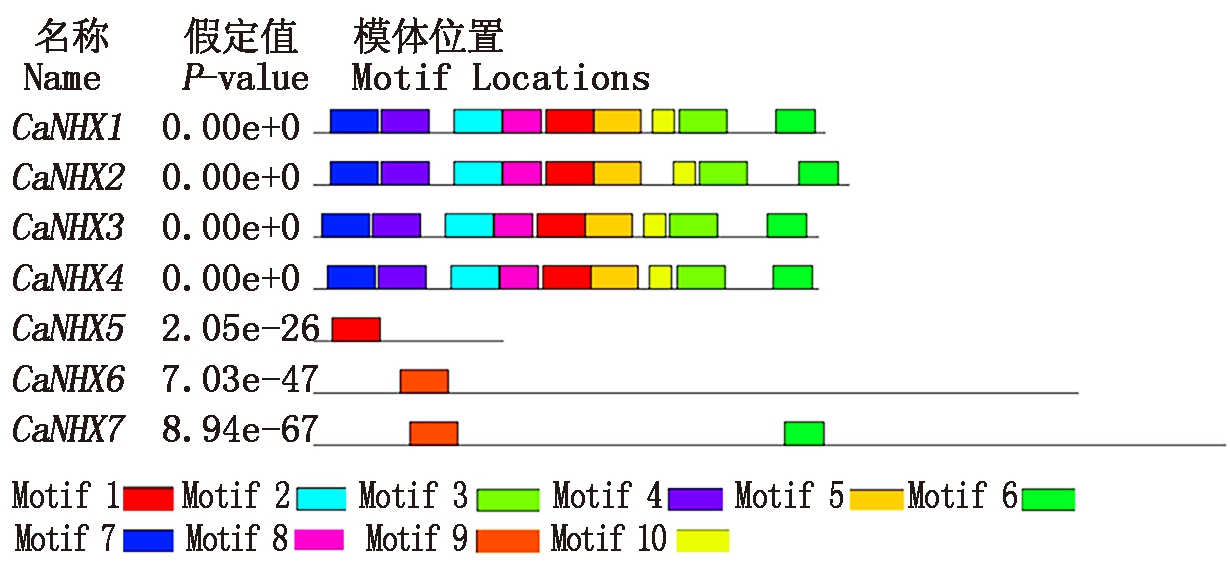
图1 CaNHXs基因家族motif分析
Fig.1 CaNHXs gene family motif analysis
2.4 CaNHXs基因家族基因结构分析
CaNHXs基因家族的结构框架通过FGENESH-C分析表明(图3),CaNHXs基因家族成员均含有上下游完整基因框架序列。CaNHXs基因家族成员不仅在基因长度上存在差异,而且在内含子和外显子数量上也差异明显,但其基因结构相似。CaNHX1~ CaNHX4基因中均含有14个外显子和13个内含子,CaNHX5、 CaNHX6和 CaNHX7基因分别含有7,19和21个外显子以及6,18,20个内含子。
2.5 CaNHXs蛋白二级结构对比分析
对辣椒NHX蛋白二级结构通过Prabi在线结果表明(表4),7个NHX蛋白质二级结构完整,主要以不规则卷曲 (Random coil)和α-螺旋 (Alpha helix)为主。

图2 MEME预测的保守位点LOCO
Fig.2 LOCO predicted by MEME
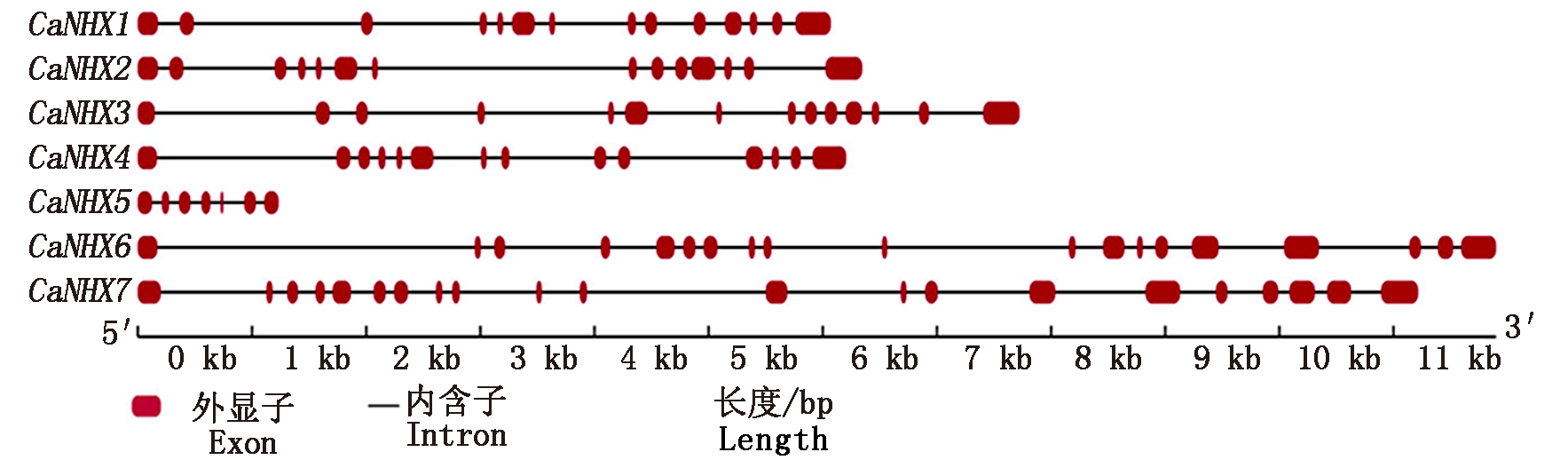
图3 CaNHXs基因结构
Fig.3 Gene structures of CaNHXs
表4 CaNHXs蛋白二级结构对比分析
Tab.4 The secondary structure of CaNHXs protein sequence %
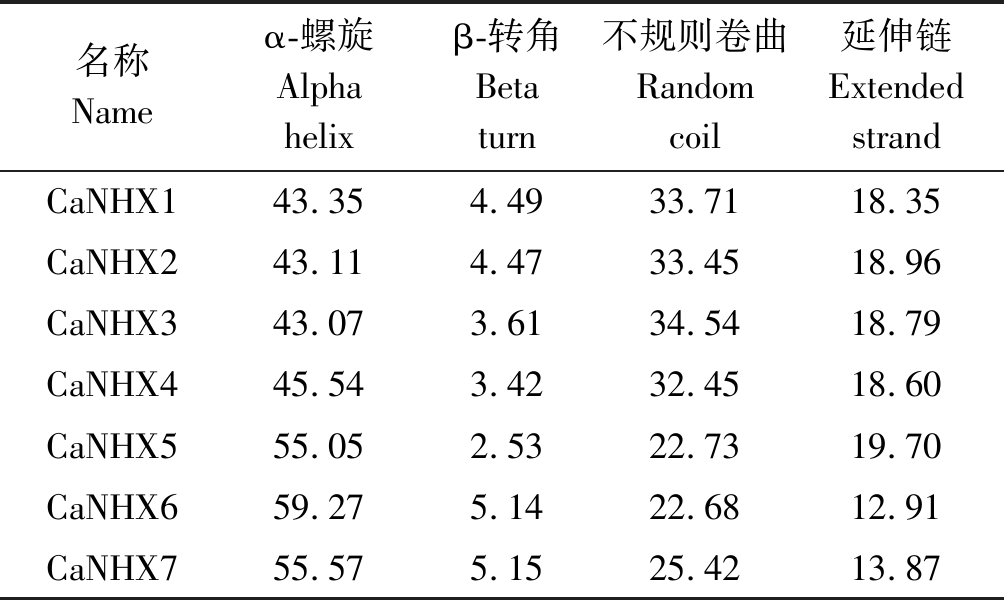
名称Name α-螺旋Alpha helix β-转角Beta turn 不规则卷曲Random coil延伸链Extended strand CaNHX143.354.4933.7118.35CaNHX243.114.4733.4518.96CaNHX343.073.6134.5418.79CaNHX445.543.4232.4518.60CaNHX555.052.5322.7319.70CaNHX659.275.1422.6812.91CaNHX755.575.1525.4213.87
2.6 CaNHXs基因顺式作用元件分析
使用PlantCare软件对辣椒NHX基因启动子序列的相关顺式调控元件进行分析发现(表5),7条NHX具有的作用元件大致分为植物激素应答元件,如茉莉酸应答元件(TGACG)等。植物生长发育相关顺式作用元件,如植物分生组织表达应答元件(CAT-box)等。生物和非生物胁迫响应顺式作用元件,如冷胁迫应答元件(LTR)、光响应元件(G-box)、盐胁迫应答元件(DRE)等。结果表明,CaNHXs在植株细胞分裂、激素信号响应以及在逆境生长中有调节作用。
2.7 CaNHXs蛋白序列系统发育树分析
为分析CaNHXs基因家族的进化,以拟南芥、番茄、胡杨、水稻NHX家族基因做参考。通过MEGA 5.0和ClustalX建立蛋白序列系统发育树。结果表明(图4),依据亲缘关系该基因家族可分为3个分支,其中AtNHX3、CaNHX4、AtNHX1、PeNHX4、AtNHX8、CaNHX5、OsNHX5、PeNHX5、CaNHX7、AtNHX7、PeNHX6为一支,CaNHX3、OsNHX4、CaNHX6、PeNHX1、SlNHX2、AtNHX5、AtNHX6、OsNHX2为一支,剩余的为一支。
2.8 CaNHXs蛋白三级结构分析
通过在线软件phyre对CaNHXs家族蛋白进行预测发现(图5),除CaNHX1~CaNHX4的蛋白三级结构高度保持一致,剩余CaNHX5、CaNHX6 和CaNHX7的蛋白三级结构各不相同。CaNHXs家族蛋白三级结构的不同是由于其基因结构中内含子/外显子之间数目和分配位置大小差距过大,进而致使各个基因组和编码氨基酸数目上存在较大的差异水平;其次所编码的蛋白二级结构中α-螺旋、β-转角、延伸链、不规则卷曲的不同导致空间折叠的不同。
表5 CaNHXs家族成员启动子序列顺式作用元件分析
Tab.5 Analysis of cis-acting elements of promoter sequence of CaNHXs family members
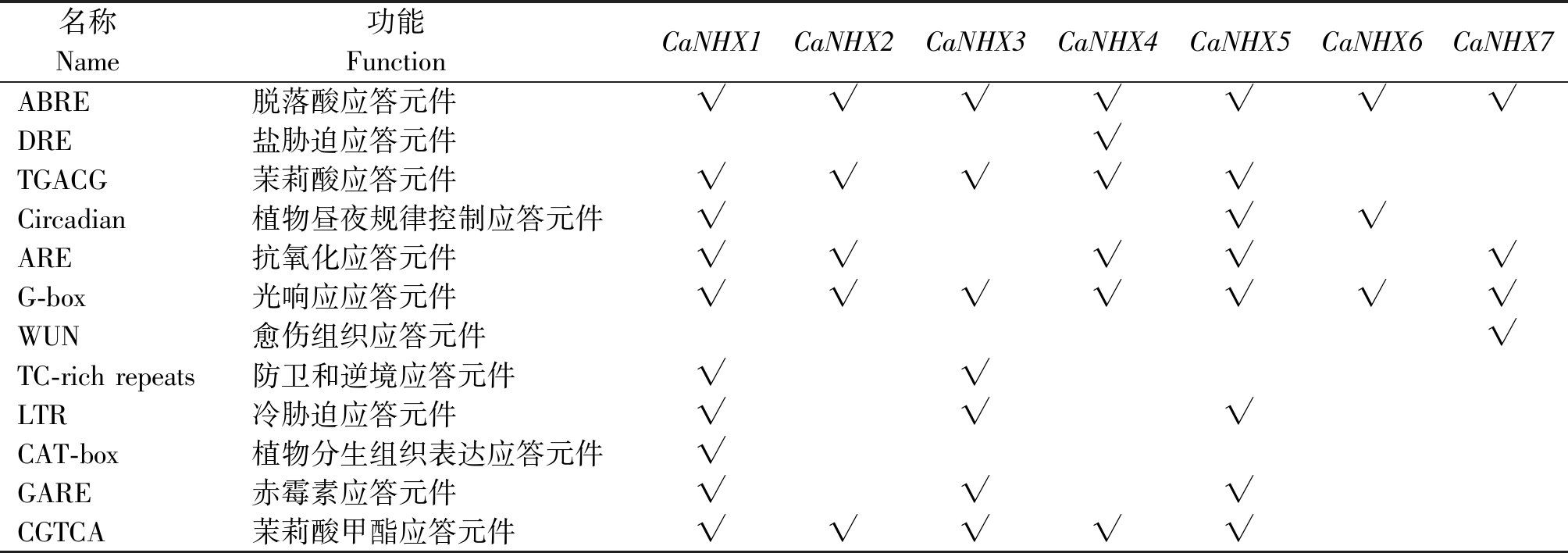
名称 Name 功能 Function CaNHX1CaNHX2CaNHX3CaNHX4CaNHX5CaNHX6CaNHX7ABRE脱落酸应答元件√√√√√√√DRE盐胁迫应答元件√TGACG茉莉酸应答元件√√√√√Circadian 植物昼夜规律控制应答元件√√√ARE抗氧化应答元件√√√√√G-box光响应应答元件√√√√√√√WUN愈伤组织应答元件√TC-rich repeats防卫和逆境应答元件√√LTR冷胁迫应答元件√√√CAT-box植物分生组织表达应答元件√GARE赤霉素应答元件√√√CGTCA茉莉酸甲酯应答元件√√√√√
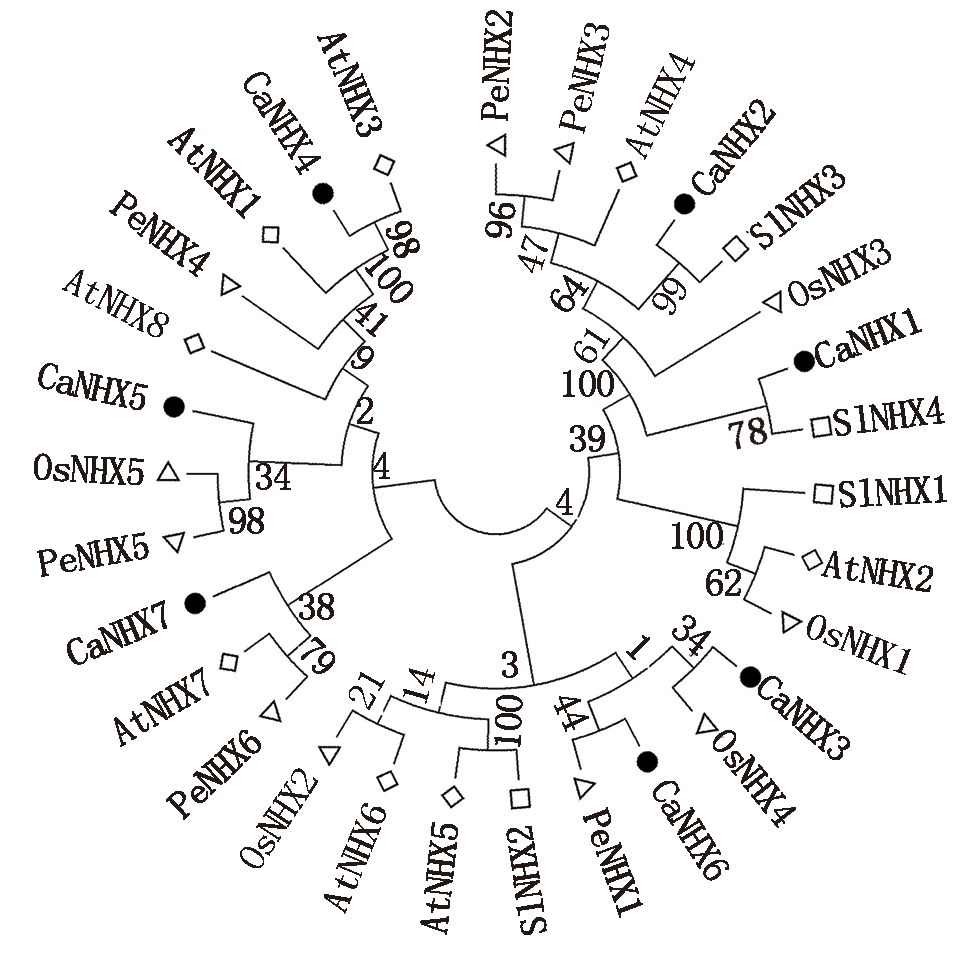
Ca.辣椒;At.拟南芥:Os.水稻;Pe.胡杨;Sl.番茄。
Ca.Capsicum annuum L.;At.Arabidopsis thaliana;Os.Oryza sativa L.;
Pe.Populus euphratica;Sl.Solanum lycopersicum.
图4 多物种NHX家族系统发育树
Fig.4 Multi-species NHX family phylogenetic tree
2.9 CaNHXs基因家族的表达分析
通过定量分析了CaNHXs家族基因在非生物胁迫差异处理下的表达情况(图6),包括盐胁迫 (200 μmol/L)、茉莉酸甲酯 (200 μmol/L)、赤霉素(100 μmol/L)和渗透胁迫(PEG 5%浓度甘露醇)。CaNHX1在GA3胁迫下,表达量下调表达在6 h时出现最小值,较0 h相比降低了28%,差异显著;在PEG胁迫下,表达量显著上调在6 h出现最大峰值,为0 h的1.8倍;而在JA胁迫下,表达量下调表达,茉莉酸甲酯处理完全抑制了CaNHX1在叶片中的活性,其表达量在12 h出现最小值,较0 h相比降低了93%,差异显著;在NaCl胁迫下,表达量显著上调表达在12 h出现最大值,为0 h的2.8倍。CaNHX2在GA3胁迫下,表达量上调表达,在12 h出现最大值,其表达量是0 h的2.3倍,差异显著;在PEG胁迫下,其表达量在6 h出现峰值,较0 h增加了50%;在JA处理下,表达量下调,在6 h出现最小值,较0 h减少了82%,差异显著;在NaCl胁迫下,表达量显著上调,在12 h出现峰值,为0 h的6倍。CaNHX3在GA3胁迫下,表达量显著上调,在12 h出现最大值,为0 h的1.8倍;在PEG胁迫下,其表达量上调表达,在6 h出现峰值,为0 h的1.5倍;在JA处理下,表达量上调表达,在12 h出现最大值,为0 h的1.8倍;在NaCl胁迫下,表达量显著下调,在24 h出现最小值,较0 h减少了84%。CaNHX4在GA3胁迫下,表达量下调表达在12 h出现最小值,较0 h相比降低了31%;在PEG胁迫下,表达量显著上调,在12 h出现最大峰值,为0 h的2.4倍;而在JA胁迫下,表达量显著下调,其表达量在12 h出现最小值,较0 h相比降低了23%;在NaCl胁迫下,表达量显著上调,在6 h出现最大值,为0 h的2.5倍。CaNHX5在GA3胁迫下,表达量显著上调,在6 h出现最大值,为0 h的2.6倍;在PEG胁迫下,表达量下调在12 h出现最小值,较0 h减少了28%;在JA胁迫下,表达量上调,其表达量在6 h出现最大值,较0 h相比增加了52%,且差异显著;在NaCl胁迫下,表达量上调,在6 h出现最大值,较0 h相比增加了13%。CaNHX6在GA3胁迫下,表达量上调,在6 h出现最大值,为0 h的4.5倍;在PEG胁迫下,表达量显著上调,在12 h出现最大峰值,为0 h的13.5倍;在JA胁迫下,表达量上调,其表达量在6 h出现最大值,较0 h相比增加了37%;在NaCl胁迫下,表达量上调,在6 h出现最大值,为0 h的13.6倍。CaNHX7在GA3胁迫下,表达量下调,在12 h出现最小值,相比0 h降低了29%;在PEG胁迫下,表达量上调,在6 h出现最大峰值,为0 h的1.7倍;在JA胁迫下,表达量显著下调,其表达量在12 h出现最小值,较0 h相比降低了16%;在NaCl胁迫下,表达量上调表达在12 h出现最大值,为0 h的2.7倍,且差异显著。
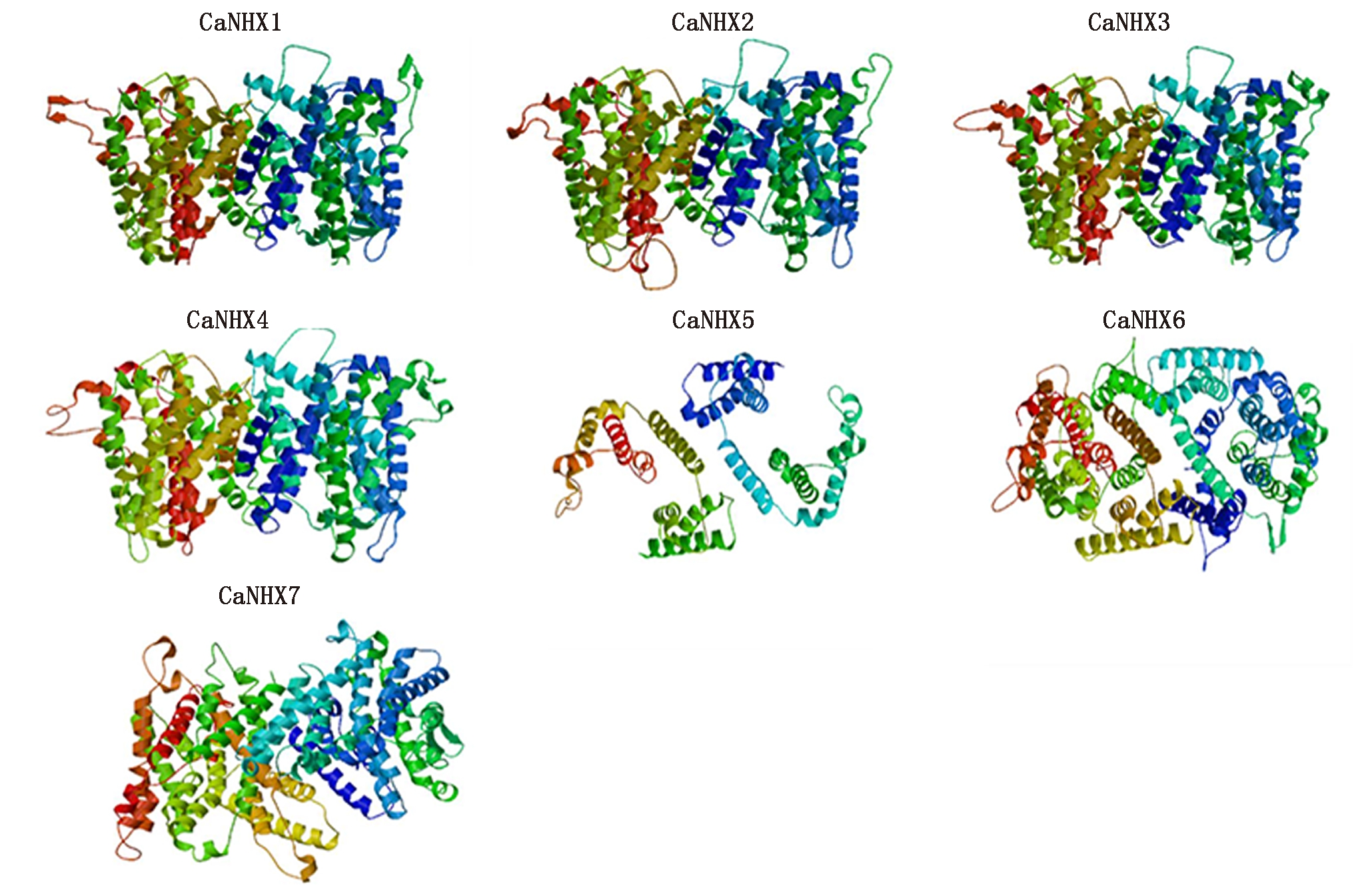
图5 CaNHXs蛋白质三级结构
Fig.5 Tertiary structure of predicted CaNHXs proteins
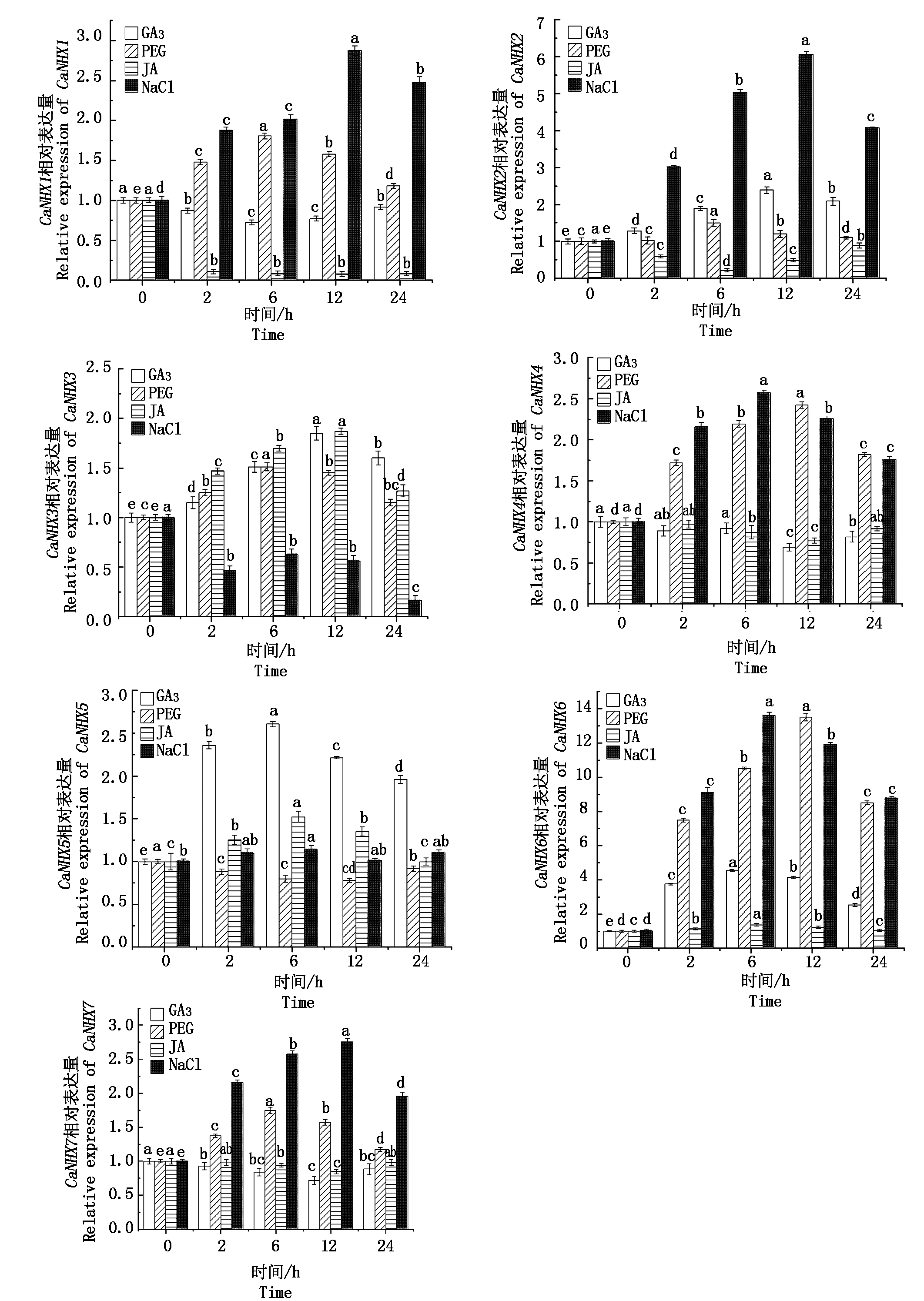
图中不同字母表示处理之间差异显著(P<0.05)。
Different letters in the same group indicate significant differences between treatments(P<0.05).
图6 CaNHXs基因对胁迫响应的表达
Fig.6 Expression of CaNHXs gene in response to stress
3 结论与讨论
目前,在陆地棉[40]、藜麦[41]、水稻[42]、葡萄[43]等植物中均有对NHX基因家族的研究;Li等[42]研究认为,植物NHX反转运蛋白对于葡萄细胞的pH值、Na+和K+稳态和盐耐受性起到重要调节作用,同时NHX的表达在一定程度上对葡萄生长开花起到影响作用。Krishnamurthy等[44]研究发现,转基因AoNHX1在拟南芥和水稻中的高表达,有利于其对土壤盐分耐受性的增加和植株在盐渍土壤中成活率升高。尽管已经在许多物种中进行了基因组和功能研究,但尚未描述辣椒NHX家族。与参考文献部分不符。
通过对辣椒中NHX基因家族生物信息学功能探究发现,其蛋白理化性质中,7个辣椒NHX基因各自分布在6条染色体上,其基因大小之间差异明显,各基因等电点变化范围差异较大,分布在5.53~9.18。CaNHX4和CaNHX7的不稳定系数均大于40,这2个基因编码产物不稳定。7个NHX蛋白中CaNHX1~CaNHX4、CaNHX6和CaNHX7均不具有信号肽,不属于分泌蛋白类型,而CaNHX5则相反。辣椒中7个NHX蛋白疏水性较高,整条多肽链表现为疏水性,其N 端含有大量的疏水氨基酸这与郭强等[45]研究结果相似,且均有明显的跨膜结构,都属于跨膜蛋白。CaNHXs基因家族成员在细胞膜(除CaNHX5)、内质网和液泡中均有着不同程度的表达。7个NHX基因序列有较高的同源性,同时均包含Na+/H+ exchange保守结构域,且在功能域上都是高度保守的,都含有上游和下游基因序列,基因结构完整。其二级结构包含内容完整。在蛋白序列的系统发育树中属于不同分支,可能这7个NHX在辣椒体内有着不同调控作用。顺式作用元件分析说明,CaNHXs在植株细胞分裂、激素信号响应以及在逆境生长中有调节作用。分析蛋白三级结构发现,除CaNHX1~CaNHX4的蛋白三级结构高度保持一致,剩余CaNHX5、CaNHX6 和CaNHX7的蛋白三级结构由于其基因结构中内含子/外显子之间数目和分配位置大小差距过大,进而致使各个基因组和编码氨基酸数目上存在较大的差异水平使得三级结构各不相同。
本试验研究表明,在NaCl处理后,CaNHX1~CaNHX7在辣椒叶片中的相对表达量均呈上调趋势,其中CaNHX6的基因表达量最高,是0 h的13.6倍,这与王影等[46]和Ohta等[47]的研究结果基本相似,郭强等[45]研究马蔺时发现,随着NaCl浓度从0~200 mmol/L 增加时,叶片中NHX基因表达量显著增大,刘雪华等[48]发现,在150 mmol/L NaCl处理下,受到盐分调节和诱导荞麦体内NHX基因表达量显著增加。在GA3处理后,CaNHX5和CaNHX6在辣椒叶片中表达量均呈显著上调趋势,在JA处理后,辣椒叶片中CaNHX1和CaNHX2以及CaNHX4和 CaNHX7的表达量受到明显抑制作用,NHX基因表达量显著降低,其余则相反。在PEG处理下,辣椒CaNHX1~CaNHX7基因表达量在叶片中均上调表达,其中CaNHX6的基因表达量最高,是0 h的13.5倍。卢世雄等[49]研究葡萄在10% PEG胁迫时发现,叶片中NHX基因表达量显著升高为对照的5倍。
总之,本研究所获得的结果,可以为进一步探索辣椒CaNHX基因的功能开发以及辣椒耐盐抗旱育种基因提供基础。
[1] Munns R,Tester M. Mechanisms of salinity tolerance[J]. Annual Review of Plant Biology,2008,59:651-681. doi:10.1146/annurev.arplant.59.032607.092911.
[2] Munns R,James R A,Xu B,Athman A,Conn S J,Jordans C,Byrt C S,Hare R A,Tyerman S D,Tester M,Plett D,Gilliham M. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene[J]. Nature Biotechnology,2012,30(3):360-364. doi:10.1038/nbt.2120.
[3] Huang X S,Wang W,Zhang Q,Liu J H. A basic helix-loop-helix transcription factor,PtrbHLH,of Poncirus trifoliata confers cold tolerance and modulates Peroxidase-mediated scavenging of hydrogen peroxide[J]. Plant Physiology,2013,162(2):1178-1194. doi:10.1104/pp.112.210740.
[4] Bassil E,Blumwald E.The ins and outs of intracellular ion homeostasis:NHX-type cation/H transporters[J].Current Opinion in Plant Biology,2014,22(22):1-6.doi:10.1016/j.pbi.2014.08.002.
[5] Zhao Q,Zhang H,Wang T,Chen S X,Dai S J. Proteomics-based investigation of salt-responsive mechanisms in plant roots[J]. Journal of Proteomices,2013,82:230-253. doi:10.1016/j.jprot.2013.01.024.
[6] Liang W J,Ma X L,Wan P,Liu L Y. Plant salt-tolerance mechanism:A review[J]. Biochemical and Biophysical Research Communications,2018,495(1):286-291. doi:10.1016/j.bbrc.2017.11.043.
[7] Apse M P,Sottosanto J B,Blumwald E. Vacuolar cation/H+ exchange,ion homeostasis,and leaf development are altered in a T-DNA insertional mutant of AtNHX1,the Arabidopsis vacuolar Na+/H+ antiporter[J]. The Plant Journal,2003,36(2):229-239. doi:10.1046/j.1365-313X.2003.01871.x.
[8] Ohnishi M,Fukada-Tanaka S,Hoshino A,Takada J,Inagaki Y,Iida S. Characterization of a novel Na+/H+ antiporter gene InNHX2 and comparison of InNHX2 with InNHX1,which is responsible for blue flower coloration by increasing the vacuolar pH in the Japanese morning glory[J]. Plant and Cell Physiology,2005 ,46(2):259-267. doi:10.1093/pcp/pci028.
[9] Bassil E,Coku A,Blumwald E. Cellular ion homeostasis:Emerging roles of intracellular NHX Na+/H + antiporters in plant growth and development[J]. Journal of Experimental Botany,2012,63(16):5727-5740.doi:10.1093/jxb/ers250.
[10] Huertas R,Rubio L,Cagnac O,Garc a-S
a-S nchez M J,Alché J D,Venema K,Fern ndez J A,Rodríguez-Rosales M P. The K+/H+ antiporter LeNHX2 increases salt tolerance by improving K + homeostasis in transgenic tomato[J]. Plant,Cell & Environment,2013,36(12):2135-2149. doi:10.1111/pce.12109.
nchez M J,Alché J D,Venema K,Fern ndez J A,Rodríguez-Rosales M P. The K+/H+ antiporter LeNHX2 increases salt tolerance by improving K + homeostasis in transgenic tomato[J]. Plant,Cell & Environment,2013,36(12):2135-2149. doi:10.1111/pce.12109.
[11] Jiang X Y,Leidi E O ,Pardo J M. How do vacuolar NHX exchangers function in plant salt tolerance? [J]. Plant Signaling & Behavior,2010,5(7):792-795. doi:10.4161/psb.5.7.11767.
[12] Qiu Q S. Plant and yeast NHX antiporters:roles in membrane trafficking[J].Journal of Integrative Plant Biology,2012,54(2):66-72. doi:10.1111/j.1744-7909.2012.01097.x.
[13] 邱全胜.拟南芥NHX5和NHX6:离子平衡与蛋白质运输[J].中国科学(生命科学),2017,47(8):839-846.doi:10.1360/N052016-00351.
Qiu Q S. Arbidopsis NHX5 and NHX6:ion homeostasis and protein transport[J].Science in China(Series C),2017,47(8):839-846.
[14] Guo Q,Meng L,Mao P C,Tian X X. Role of silicon in alleviating salt-induced toxicity in white clover[J]. Bulletin of Environmental Contamination and Toxicology,2013,91(2):213-216.doi:10.1007/s00128-013-1034-3.
[15] Blumwald E,Poole R J. Na/H antiporter in isolated tonoplast vesicles from storage tissue of Beta vulgaris[J]. Plant Physiol,1985,78:163-167. doi:10.1104/pp.78.1.163 .
[16] Apse M P,Aharon G S,Snedden W A,Blumwald E.Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis[J]. Science,1999,285(5431):1256-1258. doi:10.1126/science.285.5431.1256.
[17] Zörb C,Noll A,Karl S,Leib K,Yan F,Schubert S . Molecular characterization of Na+/H+ antiporters (ZmNHX) of maize (Zea mays L.) and their expression under salt stress[J]. Journal of Plant Physiol,2005,162(1):55-66. doi:10.1016/j.jplph.2004.03.010.
[18] Ma X L,Zhang Q,Shi H Z,Zhu J K,Zhao Y X,Ma C L,Zhang H. Molecular cloning and different expression of a vacuolar Na+/H+ antiporter gene in Suae da salsa under salt stress[J]. Biologia Plantarum,2004,48(2):219-225. doi:10.1023/B:BIOP.0000033448.96998.44.
[19] Liang M X,Lin M,Lin Z Y,Zhao L,Zhao G M ,Li Q,Yin X Z. Identification,functional characterization,and expression pattern of a NaCl-inducible vacuolar Na+/H+ antiporter in chicory(Cichorium intybus L.)[J]. Plant Growth Regulation,2015,75(3):605-614.doi:10.1007/s10725-014-9963-3.
[20] Mishra S,Alavilli H,Lee B,Panda S K,Sahoo L. Cloning and characterization of a novel vacuolar Na+/H+ antiporter gene (VuNHX1) from drought hardy legume,cowpea for salt tolerance[J]. Plant Cell,Tissue and Organ Culture,2015,120 (1):19-33.doi:10.1007/s11240-014-0572-7.
[21] Wu C A,Yang G D,Meng Q W,Zheng C C. The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress[J]. Plant and Cell Physiology,2004,45(5):600-607. doi:10.1093/pcp/pch071.
[22] Gaxiola R A,Rao R,Sherman A,Grisafi P,Alper S L,Fink G R. The Arabidopsis thaliana proton transporters,AtNhx1 and Avp1,can function in cation detoxification in yeast[J]. PNAS,1999,96(4):1480-1485. doi:10.1073/pnas.96.4.1480.
[23] Fukuda A,Nakamura A,Tanaka Y. Molecular cloning and expression of the Na+/H+exchanger gene in Oryza sativa[J]. Biochim Biophys Acta,1999,1446(1/2):149-155. doi:10.1016/S0167-4781(99)00065-2.
[24] 郭会敏,顾春笋,陈发棣,徐迎春,刘兆磊.荷花NnNHX1基因耐盐性在转化烟草中的验证[J].园艺学报,2012,39(2):323-332. doi:10.16420/j.issn.0513-353x.2012.02.005.
Guo H M,Gu C S,Chen F D,Xu Y C,Liu Z L. Salt tolerance verification of Lotus NnNHX1 in transformed tobacco[J]. Acta Horticulturae Sinica,2012,39(2):323-332.
[25] Zhang H X,Hodson J N,Williams J P,Blumwald E. Engineering salt-tolerant Brassica plants:Characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation[J].PNAS,2001,98(22):12832-12836. doi:10.1073/pnas.231476498.
[26] He C X,Yan J Q,Shen G X,Fu L H,Holaday A S,Auld D,Blumwald E,Zhang H. Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field[J]. Plant and Cell Physiology,2005,46(11):1848-1854. doi:10.1093/pcp/pci201.
[27] Bao A K,Wang Y W,Xi J J,Liu C,Zhang J L,Wang S M. Coexpression of xerophyte Zygophyllum xanthoxylum ZxNHX and ZxVP1-1 enhances salt and drought tolerance in transgenic Lotus corniculatus by increasing cations accumulation[J]. Functional Plant Biology,2014,41(2):203-214. doi:10.1071/FP13106.
[28] Su Q,Zheng X,Tian Y,Wang C. Exogenous brassinolide alleviates salt stress in Malus hupehensis Rehd. by regulating the transcription of NHX-Type Na+(K+)/H+ antiporters[J]. Frontiers in Plant Science,2020,11:38. doi:10.3389/fpls.2020.00038.
[29] Zhang H X,Blumwald E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit[J]. Natural Biotechnology,2001,19(8):765-768. doi:10.1038/90824.
[30] Jan A T,Singhal P,Haq Q M R. Plant abiotic stress:deciphering remedial strategies for emerging problem[J]. Journal of Plant Interactions,2013,8(2):97-108. doi:10.1080/17429145.2012.702226.
[31] Long L,Zhao J R,Guo D D,Ma X N,Xu F C,Yang W W,Gao W. Identification of NHXs in Gossypium species and the positive role of GhNHX1 in salt tolerance[J]. BMC Plant Biology,2020 ,20(1):147. doi:10.21203/rs.2.10038/v3.
[32] Mushke R, Rajesh Y,Kirti P B. Improved salinity tolerance and growth performance in transgenic sunflower plants via ectopic expression of a wheat antiporter gene (TaNHX2)[J]. Molecular Biology Reports,2019,46(6):5941-5953. doi:10.1007/s11033-019-05028-7.
[33] Wu X X,Li J,Wu X D,Liu Q,Wang Z K,Liu S S,Li S N,Ma Y L,Sun J,Zhao L,Li H Y,Li D M,Li W B,Su A Y. Ectopic expression of Arabidopsis thaliana Na+(K+)/H+ antiporter gene,AtNHX5,enhances soybean salt tolerance[J]. Genetics and Molecular Research,2016,15(2):1-12. doi:10.4238/gmr.15027483.
[34] 罗建,张国斌,车旭升,秦启杰.辣椒抗坏血酸生物合成酶GME基因家族的鉴定和表达分析[J].华北农学报,2020,35(4):71-78. doi:10.7668/hbnxb.20190723.
Luo J,Zhang G B,Che X S,Qin Q J.Bioinformation analysis and expression analysis of GME genes family in ascorbic acid biosynthesis of pepper[J]. Acta Agriculturae Boreali-Sinica,2020,35(4):71-78.
[35] Bickmore W A,Sutherland H G E.Addressing protein localization within the nucleus[J].The European Molecular Biology Organization Journal,2002,21(6):1248-1254. doi: 10.1093/emboj/21.6.1248.
[36] 丛郁,杨顺瑛,宋志忠,郝东利,苏彦华.葡萄AMT基因家族生物信息学分析[J].中国农学通报,2011,27(25):193-199.
Cong Y,Yang S Y,Song Z Z,Hao D L,Su Y H. Bioinformatics analysis of AMT protein family in grape[J]. Chinese Agricultural Science Bulletin,2011,27(25):193-199.
[37] 刘炳臣,王茜,王跃进,张朝红.葡萄活性赤霉素合成关键基因VvGA3ox家族的克隆和表达分析[J].园艺学报,2016,43(12):2293-2303. doi:10.16420/j.issn.0513-353x.2016-0294.
Liu B C,Wang Q,Wang Y J,Zhang C H. Cloning and expression analysis of the key genes family VvGA3ox in active gibberellin synthesis of grapevine[J]. Acta Horticulturae Sinica,2016,43(12):2293-2303.
[38] Tamura K,Dudley J,Nei M,Kumar S.MEGA4:Molecular evolutionary genetics analysis (MEGA) software version 4.0[J].Molecular Biology and Evolution,2007,24 (8):1596-1599. doi: 10.1093/molbev/msm092.
[39] Chenna R,Sugawara H,Koike T,Lopez R,Gibson T J,Higgins D G,Thompson J D.Multiple sequence alignment with the Clustal series of programs[J].Nucleic Acids Research,2003,31(13):3497-3500. doi:10.1093/nar/gkg500.
[40] Ma W Y,Ren Z Y,Zhou Y,Zhao J J,Zhang F,Feng J P,Liu W,Ma X F. Genome-wide identification of the Gossypium hirsutum NHX genes reveals that the endosomal-type GhNHX4A is critical for the salt tolerance of cotton[J]. International Journal of Molecular Sciences,2020,21(20):7712. doi:10.3390/ijms21207712 .
[41] 王宇,贾冰晨,吴筱林,张东亮,褚晶,田晓芹,陈世华,郭善利.藜麦CqNHX基因家族鉴定及表达模式分析[J].烟台大学学报(自然科学与工程版),2020,33(3):270-275. doi:10.13951/j.cnki.37-1213/n.191210.
Wang Y,Jia B C,Wu X L,Zhang D L,Chu J,Tian X Q,Chen S H,Guo S L. Genome-wide identification and expression pattern analysis of NHX gene family in Chenopodium quinoa[J]. Journal of Yantai University(Natural Science and Engineering Edition),2020,33(3):270-275.
[42] Li W H,Du J,Feng H M,Wu Q,Xu G H,Shabala S,Yu L. Function of NHX-type transporters in improving rice tolerance to aluminum stress and soil acidity[J]. Planta,2020,251(4):71. doi:10.1007/s00425-020-03361-x.
[43] Ayadi M, Martins V, Ben Ayed R, Jbir R, Feki M, Mzid R, Géros H, Aifa S M, Hanana M. Genome wide identification,molecular characterization,and gene expression analyses of grapevine NHX antiporters suggest their involvement in growth,ripening,seed dormancy,and stress response[J]. Biochemical Genetics,2020,58(1):102-128. doi:10.1007/s10528-019-09930-4.
[44] Krishnamurthy P,Vishal B,Khoo K,Rajappa S,Loh C,Kumar P P. Expression of AoNHX1 increases salt tolerance of rice and Arabidopsis,and bHLH transcription factors regulate AtNHX1 and AtNHX6 in Arabidopsis[J]. Plant Cell Reports,2019,38(10):1299-1315. doi:10.1007/s00299-019-02450-w.
[45] 郭强,孟林,李杉杉,张琳,毛培春,田小霞.马蔺NHX基因的克隆与基因表达分析[J].植物生理学报,2015,51(11):2006-2012. doi:10.13592/j.cnki.ppj.2015.0472.
Guo Q,Meng L,Li S S,Zhang L,Mao P C,Tian X X. Cloning of Iris lactea var. chinensis NHX and analysis of gene expression[J]. Plant Physiology Journal,2015,51(11):2006-2012.
[46] 王影,李慧,蔺经,杨青松,张绍铃,常有宏.杜梨NHX基因家族的鉴定及其在非生物胁迫下的表达分析[J].果树学报,2019,36(7):825-836. doi:10.13925/j.cnki.gsxb.20180347.
Wang Y,Li H,Lin J,Yang Q S,Zhang S L,Chang Y H. Identification of NHX gene family in Pyrus betulaefolia and its expression under abiotic stress[J]. Journal of Fruit Science,2019,36(7):825-836.
[47] Ohta M,Hayashi Y,Nakashima A,Hamada A,Tanaka A,Nakamura T,Hayakawa T. Introduction of a Na+/H+ antiporter gene from Atriplex gmelini confers salt tolerance to rice[J]. FEBS Letters,2002,532(3):279-282. doi:10.1016/S0014-5793(02)03679-7.
[48] 刘雪华,宋琎楠,张玉喜,侯丽霞,于延冲,赵方贵,刘春英,董春海,杨洪兵.苦荞麦FtNHX1基因的克隆及表达分析[J].华北农学报,2017,32(4):49-54. doi:10.7668/hbnxb.2017.04.008.
Liu X H,Song J N,Zhang Y X,Hou L X,Yu Y C,Zhao F G,Liu C Y,Dong C H,Yang H B. Cloning and expression analysis of FtNHX1 in tartary buckwheat[J]. Acta Agriculturae Boreali-Sinica,2017,32(4):49-54.
[49] 卢世雄,许春苗,何红红,梁国平,王萍,陈佰鸿,毛娟.葡萄NHX基因家族的鉴定和表达分析[J].果树学报,2019,36(3):266-276. doi:10.13925/j.cnki.gsxb.20180361.
Lu S X,Xu C M,He H H,Liang G P,Wang P,Chen B H,Mao J. Identification and expression analysis of NHX genes family in grape[J]. Journal of Fruit Science,2019,36(3):266-276.