棉花是世界上最重要的天然纤维作物,保证棉纤维的产量对纺织工业的稳定发展具有重要意义,高产稳产始终是棉花育种工作的重要目标[1]。陆地棉在棉纤维年产量中占比95%以上[2]。因此,研究陆地棉产量相关性状的分子遗传机制,有助于进一步培育高稳产棉花新品种。
单铃质量(Boll weight,BW)、衣分(Lint percentage,LP)、子指(Seed index,SI)和果枝数(Fruit branch number,FBN)等是棉花产量的重要数量性状,且性状之间存在复杂的相互影响[3-4]。因此,QTL定位是进行产量性状分子遗传机制挖掘的重要手段。Shen等[5]以陆地棉重组自交系为试验材料,基于一张包含110个SSR标记、总图距为810.07 cM的遗传图谱,定位到11个产量相关QTL;随后通过加密图谱标记,构建了一张包含156个SSR标记、总图距1 024.4 cM的遗传图谱,定位到26个QTL[6]。Wang等[7]以海岛棉F2群体为试验材料,构建一张包含337个位点、图距2 108.34 cM的遗传图谱,定位到21个产量相关QTL位点。Yu等[8]以海陆种间回交自交系群体为试验材料,构建一张包含392个SSR标记、总图距2 895 cM的遗传图谱,定位到39个产量相关QTL。Ning等[9]以陆地棉重组自交系群体为试验材料,构建一张包含279个SSR标记、总图距1 576.25 cM的遗传图谱,定位到61个产量相关QTL。Liu等[10]以陆地棉重组自交系为试验材料,通过构建一张包含2 051个位点、总图距3 508.29 cM的遗传图谱,定位到37个产量相关QTL。由于缺乏参考基因组信息、SSR等传统分子标记多态性较差、图谱构建过程费时费力等原因,前期的研究存在定位区间较大、染色体及基因信息不完整等不足之处。随着测序技术的进步,陆地棉参考基因组信息得以公布并不断完善[11-14]。Zhang等[15]以陆地棉重组自交系群体为试验材料,开发SLAF-SNP(Specific locus amplified fragment sequencing-Single nucleotide polymorphism)标记,构建一张包含5 521个SNP、总图距3 259.37 cM的高密度遗传图谱,定位到18个多环境稳定存在的单铃质量QTL位点,注释到344个基因。Su等[16]通过SLAF-seq技术,对355份陆地棉材料的衣分性状进行关联分析,得到12个高度关联的SNP标记,并推测基因Gh_A02G1268对衣分有重要影响。Ma等[17]对419份陆地棉材料进行重测序及关联分析,结果表明Gh_D02G0025可能是影响衣分的关键基因。Sun等[3]以10 511个SNP对719份陆地棉材料的产量性状进行关联分析,得到62个显著相关的SNP,并发现基因Gh_D03G1064和Gh_D12G2354可能对产量有重要作用。Song等[18]用CottonSNP63K芯片对276份陆地棉材料衣分性状进行关联分析,得到23个显著关联的SNP和15个QTL位点,并发现基因Gh_D05G0313和Gh_D05G1124在纤维发育阶段高调表达。由此可知,基于高质量的SNP标记和高密度遗传图谱,棉花产量相关的基因信息逐渐得到挖掘。然而,由于产量相关性状分子遗传机制的复杂性,现有的研究成果仍然不够充分,特别是对高稳产品种产量相关基因的挖掘较少。
冀丰914是由河北省农林科学院粮油作物研究所培育的高产、稳产棉花品种,于2015年通过国家黄河流域棉区审定(国审棉2015003),具有高产、稳产、丰产性好等突出特点[19]。为进一步探索高产、稳产棉花产量性状的分子遗传机制,本研究以冀丰914为母本、优质自交系冀丰817为父本构建F2和F3群体,基于GBS技术构建的包含11 488个SNP标记、总图距4 202.12 cM的高密度遗传图谱,对单铃质量、衣分、子指和果枝数等性状进行QTL定位,并对重点区域进行基因注释及候选基因筛选,以期获得更多产量相关的分子遗传信息,为新品种选育及基因功能研究提供基础。
1 材料和方法
1.1 试验材料
以自育国审品种冀丰914为母本,以优质自交系冀丰817(原系名:优系817)为父本,于2018年在河北石家庄组配杂交组合,收获F1种子,同年冬天在海南加代自交,收获F2种子。于2019年在河北石家庄播种F2,行长7.00 m,行距0.76 m,株距0.20 m,共15行、413个单株。于2020年在河北石家庄播种F3群体,行长5.00 m,行距0.76 m,株距0.20 m,3次重复。同当地大田管理。
9月底自然吐絮后,人工收获F2群体单株全株棉铃并计数,收获F3群体株行20铃,称取籽棉质量、计算单铃质量,轧花后称取皮棉和子指质量;收获前调查果枝数;F3群体性状数据为3个重复平均值。
1.2 GBS测序及遗传图谱构建
在F2群体中随机选取200个单株,采用CTAB法提取幼叶DNA[20],经质检后构建文库[21-22],选择MseⅠ和TaqαⅠ (Thermo scientific Fermentas)2种酶进行酶切,回收397~420 bp长度的酶切片段,通过Illumina HiSeqTM平台进行测序。通过BWA (Burrows-Wheeler Aligner, version 0.7.17)软件[23]对测序数据进行基因组比对,通过GATK (Genome Analysis Tool-kit, version 4.0.11.0)软件[24]检测SNP标记,通过MSTMap (Minimum spanning tree map, version update 2015)软件[25]构建遗传图谱。最终构建一张包含11 488个SNP标记、总图距4 202.12 cM的遗传图谱,标记间平均距离仅有0.37 cM。
1.3 分析方法
通过Excel 2016分析数据的基本统计量,通过SPSS 21.0分析性状相关性。
通过QTLIciMapping 4.0软件分析加性QTL[26-27],分析参数为:Step=1 cM,PIN=0.001,LOD值由1 000次迭代计算确定。
基于陆地棉TM-1参考基因组[13],注释QTL位点内的基因信息;通过KOBAS 3.0进行基因的KEGG通路分析和GO富集分析[28];根据陆地棉与海岛棉的转录组信息[14]分析注释基因表达量,筛选候选基因。
2 结果与分析
2.1 表型数据的基本统计量及简单相关性分析
表1为群体性状基本统计量。可以看出,冀丰914的各性状值均大于冀丰817。分离群体中,除2020年BW外,其他性状呈现双向超亲分布,峰度和偏度的绝对值小于1,说明性状呈正态分布,适合进行QTL定位分析。由变异系数发现,子指的变异系数最大,果枝数的变异系数最小,F2群体性状的变异系数大于F3群体。
表1 亲本及群体产量性状的基本统计量
Tab.1 Basic statistics of the parents and population yield related traits

性状及环境Trait and environment冀丰914Jifeng 914冀丰817 Jifeng 817最大值 Max最小值 Min平均值 Average偏度 Skew峰度 Kurt变异数/% CV单铃质量/g2019 SJZ6.10∗∗4.906.754.666.020.840.686.89BW2020 SJZ5.93∗∗4.556.294.935.560.22-0.285.32衣分/% 2019 SJZ43.30∗∗38.7048.1636.1843.700.170.935.96LP2020 SJZ43.10∗∗41.1246.3737.9242.76-0.33-0.285.30子指/g 2019 SJZ11.50∗∗10.3012.608.4010.300.02-0.147.98SI2020 SJZ10.75∗∗8.9512.009.2010.350.16-0.526.87果枝数 FBN2020 SJZ13.10∗∗11.8013.8010.7012.340.200.155.11
注:**.在0.01水平下,亲本之间的差异达到极显著水平;SJZ.石家庄。表2-3同。
Note: ** .The difference between the parents reached the significance level of 0.01; SJZ.Shijiazhuang. The same as Tab.2-3.
表2为4个性状之间的简单相关性分析结果。可以看出,子指与单铃质量之间存在极显著的正相关关系,与衣分之间存在极显著的负相关关系。因此,实现棉花单铃质量、衣分和子指3个性状的同步提高存在较大困难。果枝数与单铃质量、衣分、子指的相关性均不显著,说明果枝的多少对单铃质量、衣分、子指3个性状的影响较小。
表2 产量性状之间的简单相关性分析
Tab.2 Correlation analysis among the yield related traits
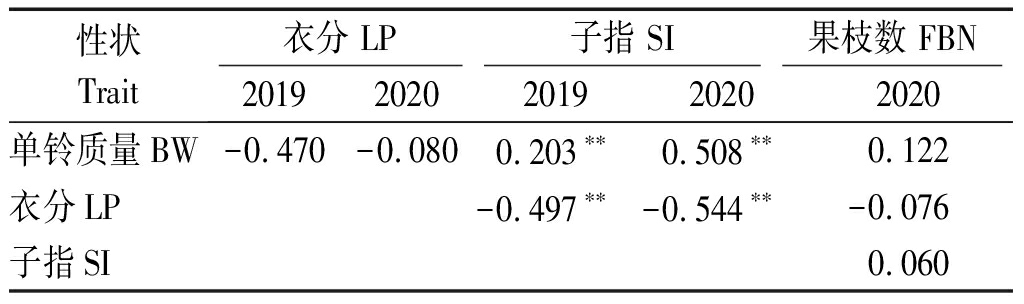
性状Trait衣分 LP子指 SI果枝数 FBN 20192020201920202020单铃质量BW-0.470-0.0800.203∗∗0.508∗∗0.122衣分LP-0.497∗∗-0.544∗∗-0.076子指SI0.060
2.2 定位到50个产量相关的QTL位点
本研究共在22条染色体上定位到50个产量相关的QTL位点,包括8个单铃质量QTL、20个衣分QTL、15个子指QTL和7个果枝数QTL(表3、图1)。8个单铃质量QTL分布于8条染色体,单个QTL对表型变异的贡献率为4.24%~9.79%,其中4个QTL的增效基因来源于冀丰914,包括qBW-A4-1、qBW-A5-1、qBW-A11-1和qBW-D10-1;qBW-A11-1可以同时在F2和F3群体中检测到;qBW-A12-1的贡献率为9.79%,增效基因来源于冀丰817。定位到20个衣分相关QTL,分布于14条染色体,其中A6、A13、D6、D10各分布有2个QTL,A7分布有3个QTL;单个QTL对表型变异的贡献率为2.82%~12.52%;冀丰914为其中15个QTL提供增效基因;qLP-A6-1能够在2个群体中检测到,贡献率分别为8.66%,4.72%,增效基因来源于冀丰914;qLP-A13-1的贡献率达到12.52%,增效基因来源于冀丰914。定位到15个子指相关QTL,分布于13条染色体,A5和D3各有2个QTL;单个QTL的贡献率为3.21%~12.96%;7个QTL的增效基因来源于冀丰914,冀丰817为另外8个QTL提供增效基因;qSI-D3-1的贡献率达到12.96%,增效基因来源于冀丰914。定位到7个果枝数QTL分布于5条染色体,D5分布有3个QTL;单个QTL的贡献率为4.00%~9.31%;冀丰914为其中2个QTL提供增效基因,冀丰817为另外5个QTL提供增效基因;qFBN-D10-1的贡献率为9.31%,增效基因来源于冀丰914。
表3 产量相关性状QTL位点信息
Tab.3 Detail information about yield related QTL

QTL名字QTL name位置/cMPosition左侧标记Left Marker右侧标记Right MarkerLOD贡献率%PVE加性效应Additive effect显性效应Dominance effect环境EnvironmentqBW-A4-111chr4_82674714chr4_825675673.145.80-0.251 7-0.375 72019SJZqBW-A5-1148chr5_2760060chr5_27594044.278.39-0.326 60.354 82019SJZqBW-A9-10chr9_78327081chr9_779875012.734.380.056 00.137 32020SJZqBW-A11-1131chr11_6087611chr11_58651873.576.24-0.104 3-0.102 52020SJZ132chr11_6087611chr11_58651873.076.29-0.098 70.817 82019SJZqBW-A12-18chr12_102749491chr12_986029934.199.790.006 10.273 62020SJZqBW-D2-184chr15_61242400chr15_612420832.815.570.331 3-0.051 02019SJZqBW-D5-1143chr18_24913700chr18_249134282.735.130.072 9-0.528 52019SJZqBW-D10-13chr23_60936408chr23_608433572.634.24-0.083 3-0.026 32020SJZqLP-A3-178chr3_87194443chr3_871697715.825.88-0.640 00.018 22020SJZqLP-A4-129chr4_80027443chr4_799976263.963.93-0.491 20.114 72020SJZqLP-A5-1107chr5_6286979chr5_62687802.743.810.537 20.564 92019SJZqLP-A6-1125chr6_14328424chr6_141976585.698.66-0.816 50.693 12019SJZ126chr6_14197658chr6_137639514.744.72-0.530 90.161 92020SJZqLP-A6-2159chr6_9132338chr6_18668174.124.360.020 9-1.135 22020SJZqLP-A7-166chr7_87412763chr7_874057374.266.37-0.181 4-1.389 62019SJZqLP-A7-1103chr7_55416466chr7_543852666.476.56-0.100 8-0.990 02020SJZqLP-A7-2147chr7_10992014chr7_109899346.826.970.637 00.145 52020SJZqLP-A9-1157chr9_3008095chr9_12892162.952.82-0.203 10.729 52020SJZqLP-A10-112chr10_112885570chr10_1128855674.657.240.836 0-0.118 02019SJZqLP-A13-1133chr13_13423372chr13_1341132911.5512.52-1.011 10.032 92020SJZqLP-A13-1143chr13_5563222chr13_55631192.974.36-0.607 00.452 32019SJZqLP-D1-1136chr14_28601539chr14_285205243.953.93-0.510 1-0.034 22020SJZqLP-D2-1128chr15_58545534chr15_585173334.366.91-0.821 30.207 22019SJZqLP-D6-139chr19_63684647chr19_636846402.723.79-0.341 00.910 12019SJZqLP-D6-149chr19_59549923chr19_595499046.646.78-0.034 01.069 92020SJZqLP-D7-199chr20_10293958chr20_92090976.286.39-0.591 70.068 32020SJZqLP-D10-145chr23_46418847chr23_450516422.573.660.029 61.025 52019SJZqLP-D10-195chr23_16971561chr23_169453012.884.03-0.144 81.067 72019SJZqLP-D13-156chr26_45730897chr26_457304663.843.79-0.447 80.166 32020SJZqSI-A3-12chr3_112439667chr3_1124361952.503.230.158 5-0.173 02019SJZqSI-A5-1127chr5_3516806chr5_35577042.694.59-0.001 0-0.304 52020SJZqSI-A5-2141chr5_2940819chr5_29003253.995.01-0.205 3-0.201 62019SJZqSI-A8-175chr8_76104161chr8_760989734.005.06-0.322 20.191 62019SJZqSI-A9-175chr9_51832653chr9_516117153.847.000.239 1-0.038 02020SJZqSI-A10-16chr10_113721207chr10_1137170484.495.71-0.214 70.136 12019SJZqSI-A13-1158chr13_1038724chr13_9857852.554.47-0.165 8-0.153 72020SJZqSI-D2-1137chr15_53033662chr15_530336504.227.680.239 30.096 52020SJZqSI-D3-154chr16_9216675chr16_66349486.9312.96-0.321 0-0.136 42020SJZqSI-D3-2102chr16_3596296chr16_35960678.0910.42-0.310 00.231 42019SJZqSI-D6-1163chr19_1074997chr19_10665244.886.310.232 4-0.099 82019SJZqSI-D7-134chr20_45720906chr20_456567343.775.110.030 1-0.412 32019SJZqSI-D8-114chr21_62555018chr21_625549185.356.920.270 2-0.036 02019SJZqSI-D11-198chr24_19582631chr24_184743972.703.210.163 6-0.147 42019SJZqSI-D12-1161chr25_655965chr25_6493402.763.570.190 1-0.041 12019SJZqFBN-A3-1111chr3_7457533chr3_74515293.725.580.056 3-0.339 42020SJZqFBN-A5-1142chr5_2940819chr5_29003255.408.26-0.220 1-0.091 52020SJZqFBN-A8-151chr8_97304255chr8_909977613.084.650.334 50.648 62020SJZqFBN-D5-18chr18_61354443chr18_613539172.894.020.122 3-0.176 02020SJZqFBN-D5-239chr18_59302916chr18_593028813.294.570.123 4-0.175 32020SJZqFBN-D5-3150chr18_14367380chr18_131065082.744.000.118 3-0.171 42020SJZqFBN-D10-196chr23_16936203chr23_168969905.659.31-0.259 3-0.013 52020SJZ
2.3 优异QTL位点的基因注释及功能分析
基于参考基因组的注释信息,本研究对qBW-A11-1、qLP-A6-1、qLP-A13-1、qSI-D3-1和qSI-D3-2等5个稳定或主效QTL进行基因信息注释。在qBW-A11-1内注释到15个基因,在qSI-D3-1内注释到26个基因,其他QTL未注释到基因信息。KEGG通路分析发现,注释基因主要参与植物细胞壁形成、纤维素生物合成、植物激素信号转导、TCA循环、代谢及氨基酸生物合成等条目。GO功能富集分析发现,共有14个显著富集的GO条目(P<0.01),包括细胞膜组分、细胞壁合成、细胞分裂素响应及多种酶活性等,共涉及10个基因;根据陆地棉TM-1和海岛棉Hai 7124的转录组信息[14],选择根、茎、叶、花、不同发育时期的纤维和胚珠等组织,对这10个基因进行表达量分析,结果发现,在所有组织中, Ghir_A11G006820.1、Ghir_D03G004880.1、Ghir_D03G004930.1和Ghir_D03G004970.1的表达量均较低,Ghir_D03G005350.1的表达量均较高,Ghir_D03G005440.1在TM-1中的表达量高于Hai 7124(图2)。

连锁群左侧数字的单位为cM。
The unit of the number on the left of linkage groups is Centi Morgan (cM).
图1 产量相关QTL位点在染色体上的分布
Fig.1 Distribution of yield related QTL on the chromosomes

A. Ghir_A11G006820.1; B. Ghir_A11G006840.1; C. Ghir_A11G006910.1; D. Ghir_D03G004880.1; E. Ghir_D03G004910.1;
F. Ghir_D03G004930.1; G. Ghir_D03G004970.1; H. Ghir_D03G005160.1; I. Ghir_D03G005350.1; J. Ghir_D03G005440.1。
图2 基因在不同组织中的表达量
Fig.2 Gene expression levels in different organs
3 讨论
产量是获得效益的基础,高产、稳产是棉花育种及生产的主要目标[1]。然而,由于植棉面积的减少,提高单产成为增加总产的重要保证,也是育种工作的重要挑战。据研究,品种改良对棉花单产增加的贡献率达到30%以上[29],而分子技术在育种方法中的应用将成为进一步提高单产的关键环节[1, 30]。因此,深入分析棉花产量及其构成因子的分子遗传机制,挖掘有效的分子标记及功能基因,是改良育种技术、提高育种效率、加快新品种培育的重要支撑[30]。本研究以高稳产国审棉花品种冀丰914为母本,通过构建F2和F3群体,结合高密度SNP遗传图谱,对产量相关的单铃质量、衣分、子指和果枝数等4个性状进行QTL定位及候选基因筛选,为进一步改良棉花产量性状奠定分子研究基础。
QTL定位是挖掘数量性状关键基因并进行综合性状分析的经典方法。Xie等[31]通过QTL定位,综合分析了小麦灌浆过程决定粒质量大小的关键因素;Zikhali等[32]在QTL定位基础上,精细定位了一个控制小麦开花的关键基因;Pan等[33]对10个玉米重组自交系的株型性状进行QTL分析,为研究玉米株型的分子遗传机制及理想株型育种奠定研究基础。在棉花中,Zhu等[34-35]精细定位了棉花鸡脚叶QTL,并得到关键候选基因GhOKRA;Xu等[36]通过经典的精细定位方法,将纤维长度QTL qFL-chr1缩小至0.9 cM区间内;Chen等[37-38]基于QTL定位结果,结合测序技术,成功得到了棉花零式果枝性状的关键基因;Ma等[39]在棉花株高相关QTL位点内得到了一个GhPIN3基因;Feng等[40]通过回交,将纤维强度QTL qFS-Chr.D02精细定位于550.66 kb范围内,得到2个候选基因。在棉花产量方面,Su等[16]在A02染色体上关联到一个衣分相关的候选基因Gh_A02G1268;Ma等[17]通过重测序技术,对产量相关的单铃质量、衣分、子指和衣指进行全基因组关联分析,得到Gh_D02G0025等候选基因;Sun等[3]分别在D03和D12染色体上关联到一个产量相关的候选基因Gh_D03G1064和Gh_D12G2354;Song等[18] 在D05染色体上关联到2个衣分相关的候选基因Gh_D05G0313和Gh_D05G1124。本研究通过开展产量相关QTL定位,得到2个稳定性较好的QTL(qBW-A11-1和qLP-A6-1)和3个贡献率大于10%的QTL(qLP-A13-1、qSI-D3-1和qSI-D3-2);基于参考基因组注释信息,通过KEGG和GO分析,在qBW-A11-1和qSI-D3-1 2个QTL内共注释到41个参与植物激素信号转导、细胞壁生物合成、氧化还原酶活性等功能的基因,其中,编码肉桂酰辅酶A还原酶2类蛋白的Ghir_D03G005440.1基因位于qSI-D3-1区段内,该基因在陆地棉TM-1多个组织中的表达量均高于Hai 7124。研究发现,肉桂酰辅酶A还原酶是木质素合成的关键酶[41-42],木质素是细胞壁形成的关键物质,在种子发育过程中有重要作用[43-44]。因此,Ghir_D03G005440.1基因可能对棉花种子发育及大小有重要影响。
本研究定位到的50个棉花产量性状QTL中,56%(28/50)位点的增效基因来源于冀丰914,其中,75%衣分相关QTL(15/20)、2个稳定QTL和3个主效QTL的增效基因均来源于冀丰914。由此说明,冀丰914对产量性状有较强的遗传控制,高衣分是其实现高稳产的重要因素。然而,由于本研究所用试验材料为F2和F3群体,研究结果需要通过高代稳定群体结合多年多点试验进行验证。
本研究通过对陆地棉产量性状进行QTL定位,获得一个在陆地棉标准系TM-1中表达量较高的Ghir_D03G005440.1基因,该基因位于子指相关的QTL区段内,可能参与棉花种子发育调控,为深入挖掘高稳产棉花品种产量性状的分子遗传机制及关键基因提供了基础;初步表明高衣分是冀丰914实现高稳产的重要因素。
[1] 喻树迅,范术丽,王寒涛,魏恒玲,庞朝友. 中国棉花高产育种研究进展[J]. 中国农业科学,2016, 49(18):3465-3476. doi: 10.3864/j.issn.0578-1752.2016.18.001.
Yu S X, Fan S L, Wang H T, Wei H L, Pang C Y. Progresses in research on cotton high yield breeding in China[J]. Scientia Agricultura Sinica,2016, 49(18):3465-3476.
[2] Chen Z J, Scheffler B E, Dennis E, Triplett B A, Zhang T Z, Guo W Z, et al. Toward sequencing cotton (Gossypium) genomes[J]. Plant Physiology,2007, 145(4):1303-1310. doi: 10.1104/pp.107.107672.
[3] Sun Z W, Wang X F, Liu Z W, Gu Q S, Zhang Y, Li Z K, et al. A genome-wide association study uncovers novel genomic regions and candidate genes of yield-related traits in upland cotton[J]. Theoretical and Applied Genetics,2018, 131(11):2413-2425. doi: 10.1007/s00122-018-3162-y.
[4] Zhang Z, Li J W, Jamshed M, Shi Y Z, Liu A Y, Gong J W, et al. Genome-wide quantitative trait loci reveal the genetic basis of cotton fibre quality and yield-related traits in a Gossypium hirsutum recombinant inbred line population[J]. Plant Biotechnology Journal,2020, 18(1):239-253. doi: 10.1111/pbi.13191.
[5] Shen X L, Zhang T Z, Guo W Z, Zhu X F, Zhang X Y. Mapping fiber and yield QTLs with main, epistatic, and QTL×environment interaction effects in recombinant inbred lines of upland cotton[J]. Crop Science,2006, 46(1):61-66. doi: 10.2135/cropsci2005.0056.
[6] Shen X L, Guo W Z, Lu Q X, Zhu X F, Yuan Y L, Zhang T Z. Genetic mapping of quantitative trait loci for fiber quality and yield trait by RIL approach in upland cotton[J]. Euphytica,2007, 155(3):371-380. doi: 10.1007/s10681-006-9338-6.
[7] Wang X Q, Yu Y, Sang J, Wu Q Z, Zhang X L, Lin Z X. Intraspecific linkage map construction and QTL mapping of yield and fiber quality of Gossypium babardense[J]. Australian Journal of Crop Science,2013, 7(9):1252-1261.
[8] Yu J W, Zhang K, Li S Y, Yu S X, Zhai H H, et al. Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum×Gossypium barbadense backcross inbred line population[J]. Theoretical and Applied Genetics,2013, 126(1):275-287. doi: 10.1007/s00122-012-1980-x.
[9] Ning Z Y, Chen H, Mei H X, Zhang T Z. Molecular tagging of QTLs for fiber quality and yield in the upland cotton cultivar Acala-Prema[J]. Euphytica,2014, 195(1):143-156. doi: 10.1007/s10681-013-0990-3.
[10] Liu X Y, Teng Z H, Wang J X, Wu T T, Zhang Z Q, et al. Enriching an intraspecific genetic map and identifying QTL for fiber quality and yield component traits across multiple environments in upland cotton (Gossypium hirsutum L.)[J]. Molecular Genetics and Genomics,2017, 292(6):1281-1306. doi: 10.1007/s00438-017-1347-8.
[11] Zhang T Z, Hu Y, Jiang W K, Fang L, Guan X Y, Chen J D, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement[J]. Nature Biotechnology,2015, 33(5):531-537. doi: 10.1038/nbt.3207.
[12] Li F G, Fan G Y, Lu C R, Xiao G H, Zou C S, Kohel R J, et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution[J]. Nature Biotechnology,2015, 33(5):524-530. doi: 10.1038/nbt.3208.
[13] Wang M J, Tu L L, Yuan D J, Zhu D, Shen C, Li J Y, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense[J]. Nature Genetics,2019, 51(2):224-229. doi: 10.1038/s41588-018-0282-x.
[14] Hu Y, Chen J D, Fang L, Zhang Z Y, Ma W, Niu Y C, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton[J]. Nature Genetics,2019, 51(4):739-748. doi: 10.1038/s41588-019-0371-5.
[15] Zhang Z, Shang H H, Shi Y Z, Huang L, Li J W, Ge Q, et al. Construction of a high-density genetic map by specific locus amplified fragment sequencing (SLAF-seq) and its application to quantitative trait loci (QTL) analysis for boll weight in upland cotton (Gossypium hirsutum)[J]. BMC Plant Biology,2016, 16(1):79. doi: 10.1186/s12870-016-0741-4.
[16] Su J J, Fan S L, Li L B, Wei H L, Wang C X, Wang H T, et al. Detection of favorable QTL alleles and candidate genes for lint percentage by GWAS in Chinese upland cotton[J]. Frontiers in Plant Science,2016, 7: 1576. doi: 10.3389/fpls.2016.01576.
[17] Ma Z Y, He S P, Wang X F, Sun J L, Zhang Y, Zhang G Y, et al. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield[J]. Nature Genetics,2018, 50(6):803-813. doi: 10.1038/s41588-018-0119-7.
[18] Song C X, Li W, Pei X Y, Liu Y G, Ren Z Y, He K L, Zhang F, Sun K, Zhou X J, Ma X F, Yang D G. Dissection of the genetic variation and candidate genes of lint percentage by a genome-wide association study in upland cotton[J]. Theoretical and Applied Genetics,2019, 132(7):1991-2002. doi: 10.1007/s00122-019-03333-0.
[19] 沈金龙,李妙,王国印,赵红霞,朱继杰,王士杰,和剑涵. 棉花新品种冀丰914在苏北地区的种植表现[J]. 中国棉花, 2017, 44(12):37-38. doi: 10.11963/1000-632X.sjqlm.20171219.
Shen J L, Li M, Wang G Y, Zhao H X, Zhu J J, Wang S J, He J H. Planting performance of new cotton cultivar Jifeng914 in Northern Jiangsu province[J]. China Cotton, 2017, 44(12):37-38.
[20] Paterson A H, Brubaker C L, Wendel J F. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis[J]. Plant Molecular Biology Reporter,1993, 11(2): 122-127. doi: 10.1007/BF02670470.
[21] Zhang Z, Wei T J, Zhong Y, Li X G, Huang J. Construction of a high-density genetic map of Ziziphus jujuba Mill. using genotyping by sequencing technology[J]. Tree Genetics & Genomes,2016,12(4):76. doi: 10.1007/s11295-016-1032-9.
[22] Zhou Z Q, Zhang C S, Zhou Y, Hao Z F, Wang Z H, et al. Genetic dissection of maize plant architecture with an ultra-high density bin map based on recombinant inbred lines[J]. BMC Genomics,2016,17(1):178. doi: 10.1186/s12864-016-2555-z.
[23] Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform[J]. Bioinformatics,2009, 25(14):1754-1760. doi: 10.1093/bioinformatics/btp324.
[24] McKenna A, Hanna M, Banks E,Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data[J]. Genome Research,2010, 20(9):1297-1303. doi: 10.1101/gr.107524.110.
[25] Wu Y H, Bhat P R, Close T J, Lonardi S. Efficient and accurate construction of genetic linkage maps from the minimum spanning tree of a graph[J]. PLoS Genetics,2008, 4(10):1-11. doi: 10.1371/journal.pgen.1000212.
[26] Li H H, Ye G Y, Wang J K. A modified algorithm for the improvement of composite interval mapping[J]. Genetics,2007, 175(1):361-374. doi: 10.1534/genetics.106.066811.
[27] Meng L, Li H H, Zhang L Y, Wang J K. QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations[J]. Crop Journal,2015, 3(3):269-283. doi: 10.1016/j.cj.2015.01.001.
[28] Xie C, Mao X Z, Huang J J, Ding Y, Wu J M, Dong S, Kong L, Gao G, Li C Y, Wei L P. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases[J]. Nucleic Acids Research,2011, 39(S2):316-322. doi: 10.1093/nar/gkr483.
[29] 孔繁玲,姜保功,张群远,杨付新,李如忠,刘永平,赵素兰,郭腾龙. 建国以来我国黄淮棉区棉花品种的遗传改良[J]. 作物学报,2000, 26(2):148-156.
Kong F L, Jiang B G, Zhang Q Y, Yang F X, Li R Z, Liu Y P, Zhao S L, Guo T L. Genetic improvements of cotton varieties in Huang-huai region in China since 1950s[J]. Acta Agronomica Sinica,2000, 26(2):148-156.
[30] Han Z G, Hu Y, Tian Q, Cao Y W, Si A J, Si Z F, et al. Genomic signatures and candidate genes of lint yield and fibre quality improvement in upland cotton in Xinjiang[J]. Plant Biotechnology Journal,2020, 18(10):2002-2014. doi: 10.1111/pbi.13356.
[31] Xie Q, Mayes S, Sparkes D L. Carpel size, grain filling, and morphology determine individual grain weight in wheat[J]. Journal of Experimental Botany,2015, 66(21):6715-6730. doi: 10.1093/jxb/erv378.
[32] Zikhali M, Wingen L U, Griffiths S. Delimitation of the Earliness per se D1 (Eps-D1) flowering gene to a subtelomeric chromosomal deletion in bread wheat (Triticum aestivum)[J]. Journal of Experimental Botany,2016, 67(1):287-299. doi: 10.1093/jxb/erv458.
[33] Pan Q C, Xu Y C, Li K, Peng Y, Zhan W, Li W Q, Li L, Yan J B. The genetic basis of plant architecture in 10 maize recombinant inbred line populations[J]. Plant Physiology, 2017, 175(2):858-873. doi: 10.1104/pp.17.00709.
[34] Zhu Q H, Spriggs A, Taylor J M, Llewellyn D, Wilson I. Transcriptome and complexity-reduced, DNA-based identification of intraspecies single-nucleotide polymorphisms in the polyploid Gossypium hirsutum L.[J]. G3-Genes, Genomes, Genetics,2014, 4(10):1893-1905. doi: 10.1534/g3.114.012542.
[35] Zhu Q H, Zhang J, Liu D X, Stiller W, Liu D J, Zhang Z S, Llewellyn D, Wilson I. Integrated mapping and characterization of the gene underlying the okra leaf trait in Gossypium hirsutum L.[J]. Journal of Experimental Botany,2016, 67(3):763-774. doi: 10.1093/jxb/erv494.
[36] Xu P, Gao J, Cao Z B, Chee P W, Guo Q, Xu Z Z, Paterson A H, Zhang X G, Shen X L. Fine mapping and candidate gene analysis of qFL-chr1, a fiber length QTL in cotton[J]. Theoretical and Applied Genetics,2017, 130(6):1309-1319. doi: 10.1007/s00122-017-2890-8.
[37] Chen W, Yao J B, Chu L, Yuan Z W, Li Y, Zhang Y S. Genetic mapping of the nulliplex-branch gene (gb_nb1) in cotton using next-generation sequencing[J]. Theoretical and Applied Genetics,2015, 128(3):539-547. doi: 10.1007/s00122-014-2452-2.
[38] Chen W, Yao J B, Li Y, Zhao L J, Liu J, Guo Y, et al. Nulliplex-branch, a TERMINAL FLOWER 1 ortholog, controls plant growth habit in cotton[J]. Theoretical and Applied Genetics,2019, 132(1):97-112. doi: 10.1007/s00122-018-3197-0.
[39] Ma J J, Pei W F, Ma Q F, Geng Y H, Liu G Y, Liu J, et al. QTL analysis and candidate gene identification for plant height in cotton based on an interspecific backcross inbred line population of Gossypium hirsutum×Gossypium barbadense[J]. Theoretical and Applied Genetics,2019, 132(9):2663-2676. doi: 10.1007/s00122-019-03380-7.
[40] Feng L C, Zhou C H, Su Q, Xu M, Yue H R, Zhang S W, Zhou B L. Fine-mapping and candidate gene analysis of qFS-Chr. D02, a QTL for fibre strength introgressed from a semi-wild cotton into Gossypium hirsutum[J]. Plant Science,2020,297:110524.doi:10.1016/j.plantsci.2020.110524.
[41] Kawasaki T, Koita H, Nakatsubo T, Hasegawa K, Wakabayashi K, Takahashi H, Umemura K, Umezawa T, Shimamoto K. Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice[J]. Proceedings of the National Academy of Sciences, 2006, 103(1):230-235. doi: 10.1073/pnas.0509875103.
[42] Pan H Y, Zhou R, Louie G V, Mühlemann J K, Bomati E K, Bowman M E, Dudareva N, Dixon R A, Noel J P, Wang X Q. Structural studies of cinnamoyl-CoA reductase and cinnamyl-alcohol dehydrogenase, key enzymes of monolignol biosynthesis[J]. The Plant Cell, 2014, 26(9):3709-3727. doi: 10.1105/tpc.114.127399.
[43] Fang L, Xu X, Li J, Zheng F, Li M Z, Yan J W, et al. Transcriptome analysis provides insights into the non-methylated lignin synthesis in Paphiopedilum armeniacum seed[J]. BMC Genomics, 2020, 21(1):524. doi: 10.1186/s12864-020-06931-1.
[44] Baldoni A, Von Pinho E V R, Fernandes J S, Abreu V M, Carvalho M L M. Gene expression in the lignin biosynthesis pathway during soybean seed development[J]. Genetics and Molecular Research, 2013, 12(3):2618-2624. doi: 10.4238/2013.February.28.2.