大麦(Hordeum vulgare L.)种植面积和产量均居世界禾谷类作物第四,仅次于玉米、水稻、小麦[1]。大麦具有适应性强、用途广泛等优点[2-3]。随着我国社会经济发展和居民食物消费结构改进,大麦的利用价值正在被人们重视[4]。株高、穗下节间长和穗长与大麦生物产量及抗倒性密切相关,生物产量是经济产量的基础和高产前提,但株高和生物产量偏高会增加大麦的倒伏风险。不同用途大麦品种的株高类性状的要求不同,青贮大麦要求株高高、穗下节间长,有利于形成高生物产量;籽粒用大麦要求株高矮、穗下节间和穗长长,有利于籽粒高产、稳产。此外,大麦株高类性状与株型构成和抗倒伏性有关。因此,株高类性状是大麦品种改良的重要目标性状[5-6]。
大麦矮化育种主要使用的半矮化基因有short culm 1 (hcm1)、semi-brachytic 1 (uzu 1)、semi-dwarf 1 (sdw1或denso)、breviarist-atum-e (ari-e)[7]。其中uzu1基因编码一个BR受体(HvBRI1)在我国大麦育种工作中得到了广泛的应用[6, 8]。株高类性状为数量性状,数量性状位点定位是研究数量性状遗传机制的重要手段[9-10]。Ren等[6]与Chen等[11]以株高、穗长、穗下各节间长为指标,在大麦7条染色体上均定位到株高、穗长、穗下节间长等性状相关的QTL。Chutimanitsakun等[12]在大麦2H的156 cM处定位到一个同时控制株高、穗长、产量的主效QTL,在1H、3H、5H、6H上定位到控制穗长的QTL。Wang等[5]在大麦7H的80 cM处定位到贡献率为23.2%的株高QTL。Peighambari等[13]筛选327个分子标记在72个大麦DH系中检测到3个株高QTL及2个穗长QTL。
随着分子标记与连锁图谱的发展,基因定位更加精确[14]。近年来,全基因组单核苷酸多态性(SNP)标记发掘与基因分型及高密度遗传图谱构建技术的发展,促进功能基因定位研究[12, 15]。本研究以我国饲用大麦泰兴9425与日本啤酒大麦Naso Nijo构建的177份DH群体及亲本为材料,构建群体的SNP分子图谱,对株高、穗下节间长和穗长进行遗传分析与QTL定位,为大麦株高类性状的分子标记辅助选择与精细定位奠定基础。
1 材料和方法
1.1 参试材料
以我国饲用大麦泰兴9425与日本啤酒大麦Naso Nijo的杂种F1的花药诱导产生的177个DH系及亲本材料。
1.2 田间设计
2018年将参试材料分别种植于新农场(32°23′N,119°23′E)和盐城市大中农场农科所试验田(33°7′N,120°39′E)。每份材料播种1行,行长1.0 m,行距0.3 m,每行人工点播20粒,3次重复。参照当地大田管理。
1.3 株高类性状调查
在参试材料收获前,每个行选取5株生长一致植株,测量株高(cm)、主穗长(cm)和穗下节间长(cm)。
1.4 DH群体遗传图谱
DH群体遗传图谱由扬州大学大麦研究所提供[16],标记分布情况见表1。该遗传图谱含有1 551个SNP标记,覆盖大麦7条染色体、分布均匀,全长957.09 cM,2个标记间的平均遗传距离为0.61 cM,且各条染色体标记间的平均距离均小于1 cM,分别为0.60,0.95,0.41,0.69,0.68,0.93,0.47 cM。
表1 DH群体遗传图谱基本信息
Tab.1 Basic information of DH population genetic map
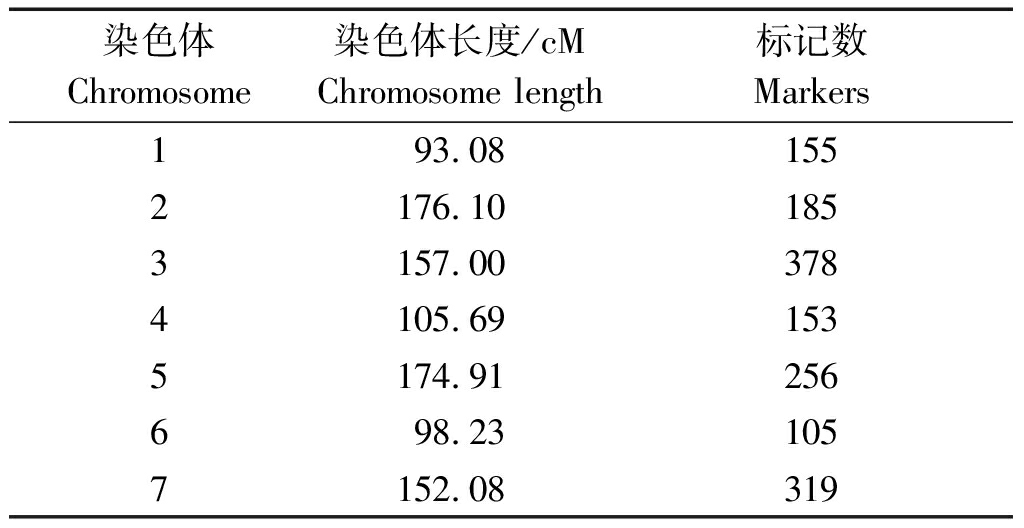
染色体Chromosome染色体长度/cMChromosome length标记数Markers193.081552176.101853157.003784105.691535174.91256698.231057152.08319
1.5 数据处理
应用Excel 2016程序对试验数据初步处理,采用SPSS 16.0软件对DH群体进行描述性统计分析、方差分析、相关性分析。利用Windows QTL IciMaping V4.1.0.0 软件(http://www.isbreeding.net/software/)对株高类性状进行QTL定位分析,LOD阈值设定为3.0。
2 结果与分析
2.1 亲本及DH群体株高类性状的表现
亲本及DH群体株高类性状表现见表2。如表2所示,泰兴9425的株高、穗长、穗下节间长均低于Naso nijo,t测验表明,株高和穗长在亲本间差异极显著。DH群体株高、穗长、穗下节间长的变异系数分别为9.52%,11.43%,10.33%。株高、穗长和穗下节间长的遗传力分别为78.98%,89.31%和84.50%。
2.2 DH群体株高类性状的方差分析
DH群体株高类性状的方差分析结果列于表3。从表3可知,3个株高类性状在DH系间的差异均达到极显著水平(P<0.01);穗长和穗下节间长在试点间差异均达到极显著水平(P<0.01);株高和穗下节间长在环境与基因型互作间差异均达极显著水平(P<0.01)。说明大麦株高类性状由基因型、试点气候及栽培条件共同决定的,穗长和穗下节间长较易受试点条件影响。
表2 亲本及DH群体株高类性状的表现
Tab.2 Plant height-related characters performance of parents and DH populations
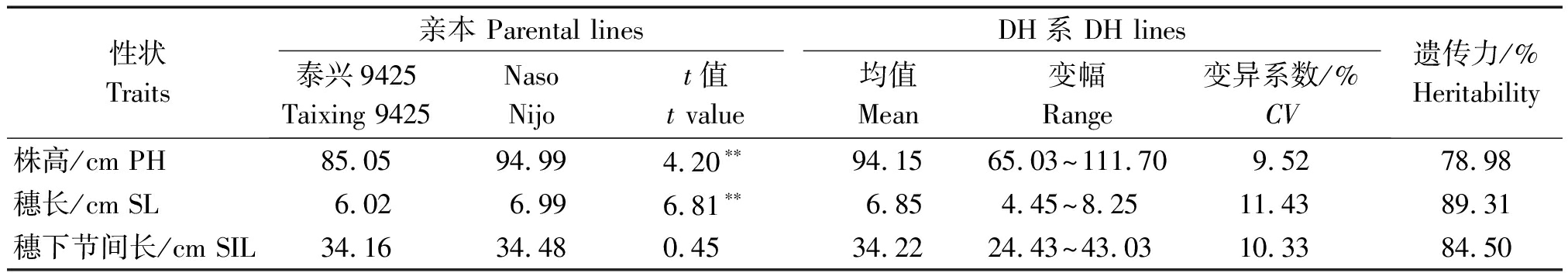
性状Traits亲本 Parental linesDH系 DH lines泰兴9425Taixing 9425Naso Nijot值 t value均值 Mean变幅 Range变异系数/%CV遗传力/%Heritability株高/cm PH85.0594.994.20∗∗94.1565.03~111.709.5278.98穗长/cm SL6.026.996.81∗∗6.854.45~8.2511.4389.31穗下节间长/cm SIL34.1634.480.4534.2224.43~43.0310.3384.50
注:*和**分别代表0.05和0.01水平上的显著。表3-4同。
Note: *and ** indicated significant difference at 0.05 and 0.01 level, respectively.The same as Tab.3-4.
表3 DH群体株高类性状的方差分析 (F值)
Tab.3 Analysis of variance on plant height-related
characters in DH population (F value)
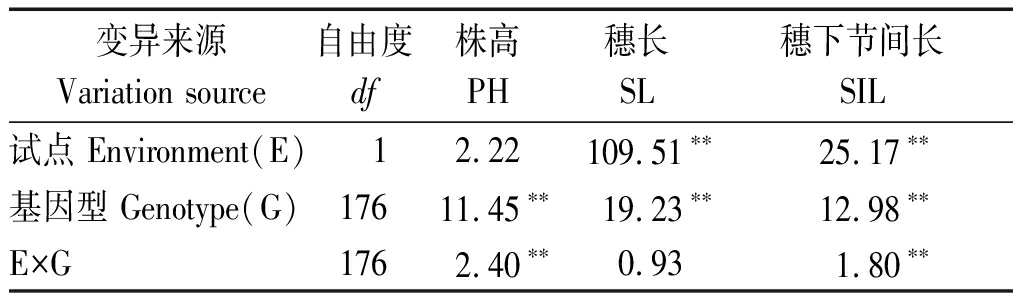
变异来源 Variation source自由度df株高 PH穗长 SL穗下节间长 SIL试点 Environment(E) 12.22109.51∗∗25.17∗∗基因型 Genotype(G)17611.45∗∗19.23∗∗12.98∗∗ E×G1762.40∗∗0.931.80∗∗
2.3 株高类性状的相关性分析
株高类性状的相关性分析结果列于表4。由表4可知,株高与穗长、株高与穗下节间长、穗长与穗下节间长均呈极显著正相关(P<0.01)。相关系数分别为0.688,0.862和0.600,表明大麦株高越高,其穗下节间长和穗长越长。
表4 株高类性状的相关性
Tab.4 Correlation coefficients between the
traits of plant height-related characters
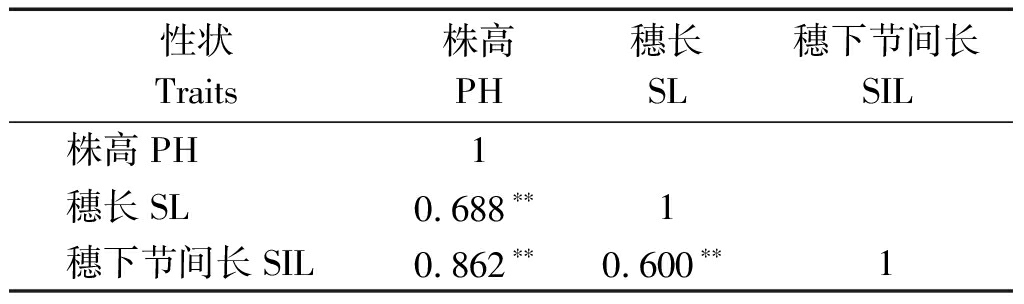
性状Traits株高PH穗长SL穗下节间长SIL株高PH1穗长SL0.688∗∗1穗下节间长SIL0.862∗∗0.600∗∗1
2.4 株高类性状的QTL分析
利用Windows QTL Icimapping V4.1.0.0软件,对3个株高类性状进行QTL定位,定位结果见表5。由表5可知,2个环境共定位到6个株高QTL,分别位于2H、3H、4H及7H上,未检测到两试点共同的QTL;两试点均值定位到4个株高QTL,其中位于3H上的qPH-3-1、4H上的qPH-4-1,与大中农场试点的2个株高QTL重演,且qPH-3-1的贡献率较大,在19.98%~38.30%。2个环境共定位到5个穗长QTL,其中QTL位于5H上的qSL-5-1在两试点重演;两试点均值定位到3个穗长QTL,包括位于5H上的qSL-5-1,但其贡献率偏小。2个环境下共定位到4个穗下节间长QTL,位于2H上的qSIL-2-1在两试点重演;两试点均值定位到2个穗下节间长QTL,包括位于2H上的qSIL-2-1,该QTL的贡献率在7.32%~13.94%。除第1,6染色体外,其他染色体上均有分布,LOD值为3.50~32.46,对表型变异的解释率为1.53%~38.30%。其中qPH-4-1、qPH-7-2、qSL-2-2、qSIL-2-1、qSIL-3-2均未见报道,可能为新位点。
3 讨论
株高是大麦最重要的性状之一,适宜的株高是确保高产的基础,既可以保证一定的生物产量,也可提高大麦的抗倒性,降低倒伏对大麦产量和品质不良影响[17]。大麦株高由包括穗长及4个左右节间与节构成,大麦穗长与穗型有关,一般垂穗型大麦品种的穗长大于直穗型品种,同一穗型品种间穗型的穗长差异较大[6, 11],穗长的长短影响到穗粒数和粒型。穗下节间长影响到植株上部功能叶的分布与通风透光性,大麦品种的穗下节长度占株高的比例越高,其产量潜力越大。目前已报道的与株高相关的QTL多受环境影响[14],利用多环境定位株高类性状的QTL,可提高定位QTL的稳定性[9, 15]。DH群体是一个永久性群体,可以实现多环境鉴定数量性状,不但可以提高定位基因的基因准确性,也可以分析基因与环境的互作效应[18]。大麦DH群体已广泛用于大麦株高类性状、产量性状、生物抗性和非生物抗性的遗传分析与QTL分析等[19-23]。本研究所用材料即为DH群体永久群体,通过多点鉴定大麦株高类性状,保证定位QTL的稳定性。
表5 株高类性状相关的QTL
Tab.5 QTL for plant height-related characters
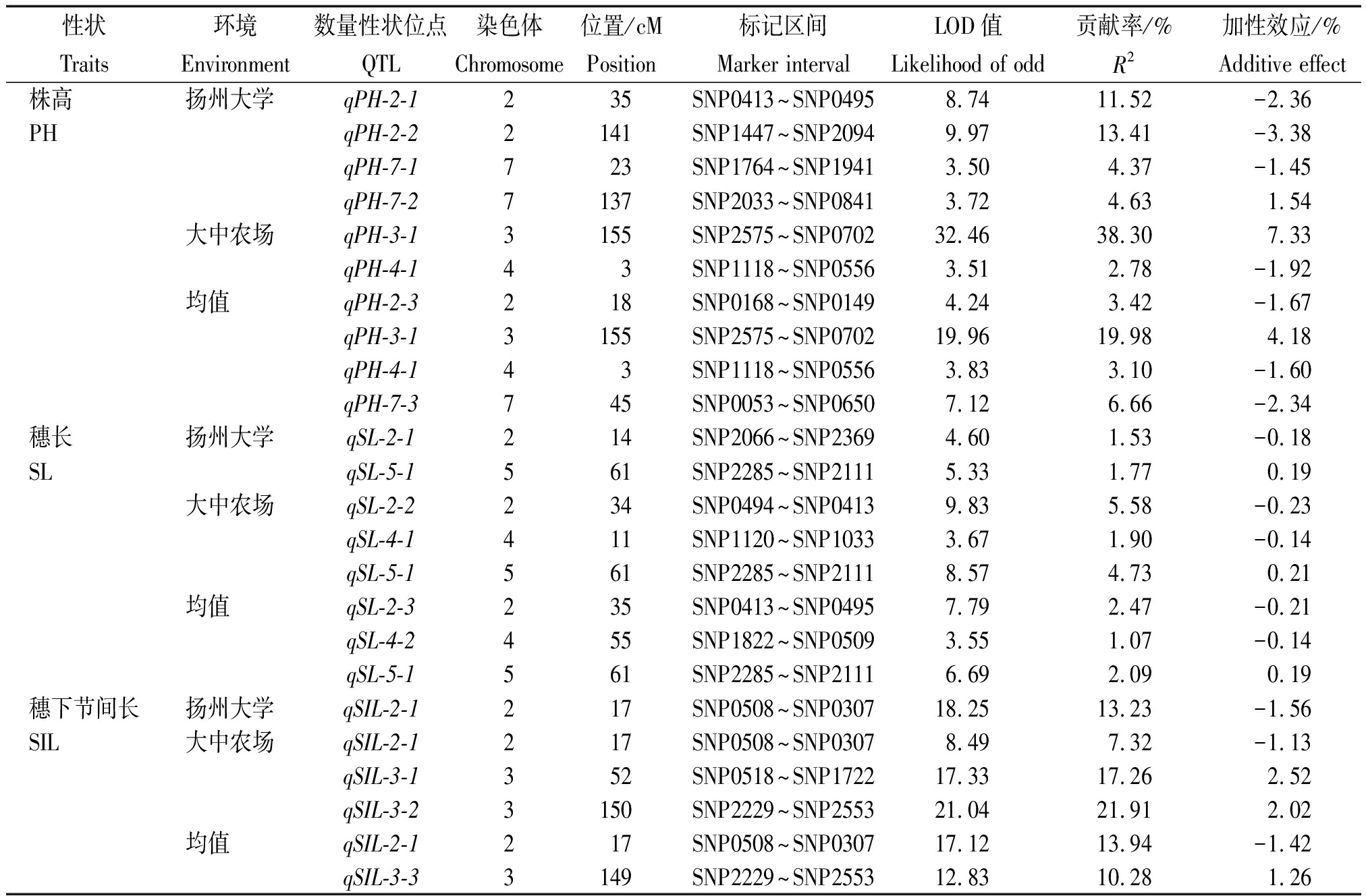
性状环境数量性状位点染色体位置/cM标记区间LOD值贡献率/%加性效应/%TraitsEnvironmentQTLChromosomePositionMarker intervalLikelihood of oddR2Additive effect株高扬州大学qPH-2-1235SNP0413~SNP04958.7411.52-2.36PHqPH-2-22141SNP1447~SNP20949.9713.41-3.38qPH-7-1723SNP1764~SNP19413.504.37-1.45qPH-7-27137SNP2033~SNP08413.724.631.54大中农场qPH-3-13155SNP2575~SNP070232.4638.307.33qPH-4-143SNP1118~SNP05563.512.78-1.92均值qPH-2-3218SNP0168~SNP01494.243.42-1.67qPH-3-13155SNP2575~SNP070219.9619.984.18qPH-4-143SNP1118~SNP05563.833.10-1.60qPH-7-3745SNP0053~SNP06507.126.66-2.34穗长扬州大学qSL-2-1214SNP2066~SNP23694.601.53-0.18SLqSL-5-1561SNP2285~SNP21115.331.770.19大中农场qSL-2-2234SNP0494~SNP04139.835.58-0.23qSL-4-1411SNP1120~SNP10333.671.90-0.14qSL-5-1561SNP2285~SNP21118.574.730.21均值qSL-2-3235SNP0413~SNP04957.792.47-0.21qSL-4-2455SNP1822~SNP05093.551.07-0.14qSL-5-1561SNP2285~SNP21116.692.090.19穗下节间长扬州大学qSIL-2-1217SNP0508~SNP030718.2513.23-1.56SIL大中农场qSIL-2-1217SNP0508~SNP03078.497.32-1.13qSIL-3-1352SNP0518~SNP172217.3317.262.52qSIL-3-23150SNP2229~SNP255321.0421.912.02均值qSIL-2-1217SNP0508~SNP030717.1213.94-1.42qSIL-3-33149SNP2229~SNP255312.8310.281.26
本研究结果表明,大麦株高、穗长、穗下节间长的遗传力较高,分别为78.98%,89.31%,84.50%,主要受遗传因素控制,可在杂交育种早代进行较为严格的选择。鉴于株高与穗长和穗下节间长间存在极显著正相关,与前人的研究相同[3, 6, 13]。因此,在对3个性状选择时,应选择株高适宜、穗下节间长占株高比例较大、穗长略长的植株。本研究共检测的6个株高相关QTL中,2H上的qPH-2-1与PH-1[7]、Qph2.1[21]、11_11505[22]距离相近,可能为同一位点。qPH-2-2、qPH-7-2分别与Yu等[7]定位到的PH-2、PH-7距离较近;3H上的qPH3-1与Schmalenbach等[23]定位的来自野生大麦导入系的QHei.S42IL-3H.a距离较近,可能为同一位点。qPH-4-1、qPH-7-2附近未报道相关位点,可能存在新的控制株高的基因。5个穗长QTL中,4H上的qSL-4-1与Islamovic等[3]定位的QTL距离较近;5H定位到qSL-5-1与Lakew等[24]定位QTL位点相同;未见与qSL-2-1、qSL-2-2位置相近的穗长QTL报道,可能为新的穗长位点。4个穗下节间长QTL中,3H上的qSIL-3-1与Ren等[6]定位到的Qion3-9距离较近,可能为同一位点;qSIL-3-2附近未报道相关QTL,可能为新的穗下节间长位点;2H上定位到的稳定的qSIL-2-1,贡献率10%左右,该QTL未报道,可能为新的穗下节间长位点,尚需进一步验证。
[1] Schulte D, Close T J, Graner A, Langridge P, Matsumoto T, Muehlbauer G, Sato K, Schulman A H, Waugh R, Wise R P,Stein N. The international barley sequencing consortium-at the threshold of efficient access to the barley genome[J].Plant Physiology, 2009, 149(1):142-147. doi:10.1104/pp.108.128967.
[2] 陈海华, 董海洲. 大麦的营养价值及在食品业中的利用[J]. 西部粮油科技, 2002, 27(2):34-36. doi: 10.3969/j.issn.1007-6395.2002.02.012.
Chen H H, Dong H Z. The nutritional value of the barley and its application in food industry[J]. China Western Cereals & Oils Technology, 2002, 27(2):34-36.
[3] Islamovic E, Obert D E, Oliver R E, Marshall J M, Miclaus K J, Hang A, Chao S, Lazo G R, Harrison S A, Ibrahim A, Jellen E N, Maughan P J, Brown R H,Jackson E W. A new genetic linkage map of barley (Hordeum vulgare L.) facilitates genetic dissection of height and spike length and angle[J].Field Crops Research, 2013, 154:91-99. doi:10.1016/j.fcr.2013.06.001.
[4] 张融, 李先德. 饲料大麦的应用价值及开发前景[J]. 中国食物与营养, 2015,21(7):29-33. doi: 10.3969/j.issn.1006-9577.2015.07.007.
Zhang R, Li X D. Application value and development prospect of feed barley in China[J]. Food and Nutrition in China, 2015,21(7):29-33.
[5] Wang J M, Yang J M, Jia Q J, Zhu J H, Shang Y, Hua W, Zhou M X. A new QTL for plant height in barley (Hordeum vulgare L.) showing no negative effects on grain yield[J].PLoS One, 2014, 9(2):e90144. doi:10.1371/journal.pone.0090144.
[6] Ren X F, Sun D F, Dong W B, Sun G L, Li C D. Molecular detection of QTL controlling plant height components in a doubled haploid barley population[J].Genetics and Molecular Research, 2014, 13(2):3089-3099. doi:10.4238/2014.April.17.5.
[7] Yu G T, Horsley R D, Zhang B X, Franckowiak J D. A new semi-dwarfing gene identified by molecular mapping of quantitative trait loci in barley[J].Theoretical and Applied Genetics, 2010, 120(4):853-861. doi:10.1007/s00122-009-1216-x.
[8] Chono M, Honda I, Zeniya H, Yoneyama K, Saisho D, Takeda K, Takatsuto S, Hoshino T, Watanabe Y. A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor[J].Plant Physiology, 2003, 133(3):1209-1219. doi:10.1104/pp.103.026195.
[9] Pasam R K, Sharma R, Malosetti M, van Eeuwijk F A, Haseneyer G, Kilian B, Graner A. Genome-wide association studies for agronomical traits in a world wide spring barley collection[J].BMC Plant Biology,2012, 12(1):16. doi:10.1186/1471-2229-12-16.
[10] Gyenis L, Yun S J, Smith K P, Steffenson B J, Bossolini E, Sanguineti M C, Muehlbauer G J. Genetic architecture of quantitative trait loci associated with morphological and agronomic trait differences in a wild by cultivated barley cross[J].Genome, 2007, 50(8):714-723. doi:10.1139/g07-054.
[11] Chen W Y, Liu Z M, Deng G B, Pan Z F, Liang J J, Zeng X Q, Tashi N M, Long H, Yu M Q. Genetic relationship between lodging and lodging components in barley (Hordeum vulgare) based on unconditional and conditional quantitative trait locus analyses[J].Genetics and Molecular Research, 2014, 13(1):1909-1925. doi:10.4238/2014.March.17.19.
[12] Chutimanitsakun Y, Nipper R W, Cuesta-Marcos A, Cistué L, Corey A, Filichkina T, Johnson E A, Hayes P M. Construction and application for QTL analysis of a restriction site associated DNA (RAD) linkage map in barley[J].BMC Genomics, 2011, 12: 4. doi:10.1186/1471-2164-12-4.
[13] Peighambari S A, Samadi B Y, Nabipour A, Charmet G, Sarrafi A. QTL analysis for agronomic traits in a barley doubled haploids population grown in Iran[J].Plant Science, 2005, 169(6):1008-1013. doi:10.1016/j.plantsci.2005.05.018.
[14] Hori K, Kobayashi T, Shimizu A, Sato K, Takeda K, Kawasaki S. Efficient construction of high-density linkage map and its application to QTL analysis in barley[J].Theoretical and Applied Genetics, 2003,107(5):806-813. doi:10.1007/s00122-003-1342-9.
[15] Ren X F, Wang J B, Liu L P, Sun G L, Li C D, Luo H, Sun D F. SNP-based high density genetic map and mapping of btwd1 dwarfing gene in barley[J].Scientific Reports, 2016, 6:31741. doi:10.1038/srep31741.
[16] Fan X Y, Zhu J, Dong W B, Sun Y D, Lü C, Guo B J, Xu R G. Comparative mapping and candidate gene analysis of SSIIa associated with grain amylopectin content in barley (Hordeum vulgare L.)[J].Frontiers in Plant Science, 2017, 8:1531. doi:10.3389/fpls.2017.01531.
[17] Day A D, Dickson A D. Effect of artificial lodging on grain and malt quality of fall-sown irrigated barley[J].Agronomy Journal, 1958, 50(6):338-340. doi:10.2134/agronj1958.00021962005000060015x.
[18] 胡中立, 何瑞锋, 章志宏, 张学富. 分子标记与QTL间连锁的检测与估计:Ⅱ.利用双单倍体(DH)群体[J]. 武汉大学学报(自然科学版), 1997, 43(6):810-814. doi: 10.1007/BF02951625.
Hu Z L, He R F, Zhang Z H, Zhang X F. Methods for detection and estimation of linkage between a marker locus and quantitative trait loci Ⅱ.using double haploid (DH) population[J]. Journal of Wuhan University(Natural Science Edition), 1997, 43(6):810-814.
[19] Obsa B T, Eglinton J, Coventry S, March T, Guillaume M, Le T P, Hayden M, Langridge P, Fleury D. Quantitative trait loci for yield and grain plumpness relative to maturity in three populations of barley (Hordeum vulgare L.) grown in a low rain-fall environment[J].PLoS One, 2017, 12(5):e0178111. doi:10.1371/journal.pone.0178111.
[20] Li J Z, Huang X Q, Heinrichs F, Ganal M W, Röder M S. Analysis of QTLs for yield, yield components, and malting quality in a BC3-DH population of spring barley[J].Theoretical and Applied Genetics, 2005, 110(2):356-363. doi:10.1007/s00122-004-1847-x.
[21] Li J Z, Huang X Q, Heinrichs F, Ganal M W, Röder M S. Analysis of QTLs for yield components, agronomic traits, and disease resistance in an advanced backcross population of spring barley[J].Genome, 2006, 49(5):454-466. doi:10.1139/g05-128.
[22] Mansour E, Casas A M, Gracia M P, Molina-Cano J L, Moralejo M, Cattivelli L, Thomas W T B, Igartua E. Quantitative trait loci for agronomic traits in an elite barley population for Mediterranean conditions[J].Molecular Breeding, 2014, 33(2):249-265. doi:10.1007/s11032-013-9946-5.
[23] Schmalenbach I, Léon J, Pillen K. Identification and verification of QTLs for agronomic traits using wild barley introgression lines[J].Theoretical and Applied Genetics, 2009, 118(3):483-497. doi:10.1007/s00122-008-0915-z.
[24] Lakew B, Henry R J, Ceccarelli S, Grando S, Eglinton J, Baum M. Genetic analysis and phenotypic associations for drought tolerance in Hordeum spontaneum introgression lines using SSR and SNP markers[J].Euphytica, 2013, 189(1):9-29. doi:10.1007/s10681-012-0674-4.