藜麦,又称南美藜、藜谷、藜米、奎奴亚藜等,属于苋科(Amaranthaceae)藜亚科(Chenopodioideae)藜属(Chenopodium),是原产于南美洲安第斯山区的一种粮食作物。藜麦被称为假谷物,虽然它不属于禾本科,但是籽粒可以像禾本科植物那样磨成面粉[1]。由于藜麦籽粒中具有丰富的营养成分,包括多种人体必需的氨基酸、含量高于其他谷物的蛋白质、脂肪、矿物质、淀粉、维生素以及异黄酮等[2-7],被联合国 FAO认定为唯一完美营养食品,有“未来的超级谷物”、“营养黄金”、“有机谷类之王”等美誉[8-9]。
藜麦对环境的适应性很广,具有较强的耐盐、抗旱、抗寒能力,在世界各大洲均有种植。根据藜麦在海拔和纬度的分布将其分为5种生态型:山谷型、高地型、盐滩型、高温湿润气候带型、海平面型。生产中高地型、盐滩型及不同类型间的杂交种较常用。藜麦具有非常丰富的表型和基因型遗传资源,尤其是籽粒颜色,多达66种,最常见的有红、白、黑3种[10]。
我国引进种植藜麦的时间较短,目前,在青海、甘肃、山西等地区大面积种植。对藜麦的研究最近一二十年才开始起步,主要集中在藜麦生理、栽培、引种试种、分子机制等方面[11-14]。在细胞学方面研究较少,尤其是核型研究相对匮乏。利用青海自己育成的品种柴达木黑-1、柴达木白-1和柴达木红-1,采用普通压片法对3个藜麦品种的核型进行研究,旨在为藜麦的系统进化、亲缘关系、基因组原位杂交等研究提供细胞学资料,并为藜麦品种选育、杂交育种、品质改良等提供科学依据。
1 材料和方法
1.1 试验材料
本试验中3个栽培藜麦品种由青海省海西州海藜(海杭)农业科技有限公司提供,分别为柴达木白-1(简称白藜麦)、柴达木黑-1(简称黑藜麦)和柴达木红-1(简称红藜麦),均为该公司自主培育的商业藜,见图1。
黑藜麦株高大于2 m,花序紧凑,呈深绿色,为复穗状花序。籽粒饱满,较大,千粒质量可达3.89 g,种子含膳食纤维及抗氧化剂最高,易于和其他谷物混合并用于冷食(调制色拉),特别适合老年人,高血压及糖尿病患者。
白藜麦植株较高,一般大于2 m,花序颜色为浅紫色,具有球形复总状花序。籽粒较大,千粒质量为3.93 g,种子口感柔和,含极高的维生素E,适合婴幼儿食用。
红藜麦植株2.5~3.0 m,花序介于紧凑和松散型,呈深紫色。籽粒千粒质量达3.54 g。种子含较高的复合维生素B,具亮丽色泽,易于和糙米混合及用于冷食。
供试种子于2018年种植在德令哈市柯鲁克镇,并于当年9月底收获备用。核型分析试验于2018年冬季至2019年春季在中国科学院高原生物适应与进化重点实验室进行。

图1 3种不同籽粒颜色藜麦品种及对应的株型
Fig.1 Three quinoa varieties with different seed color and their corresponding plant types
1.2 试验方法
中期染色体的制备:将3个藜麦品种的种子培养至根长1.0~1.5 cm,用冰水混合物在4 ℃条件下预处理24 h,乙醇∶冰醋酸(3∶1)固定至少30 min,取根尖的分生区部分,45%乙酸火焰干燥压片,用相差显微镜镜检。将一个视野下有多个清晰的中期分裂相的片子放于-80 ℃冰箱中冷冻至少30 min,经干燥后用DAPI染色,用Leica荧光显微镜观察、拍照[15]。用Photoshop cc 2015、Excel 2018进行后期的图像处理及染色体长度、臂比等的计算。根据李懋学等[16]方法统计至少30 个细胞,如果85% 以上细胞具有的恒定一致的染色体数即为该种的染色体数,然后分别选用5个分散良好的中期染色体在Image J软件中进行测量分析,得到核型数据。核型分类的依据分别采用Levan等[17]的划分标准,核型不对称系数参照Arano[18]的方法。
2 结果与分析
2.1 黑藜麦的染色体核型
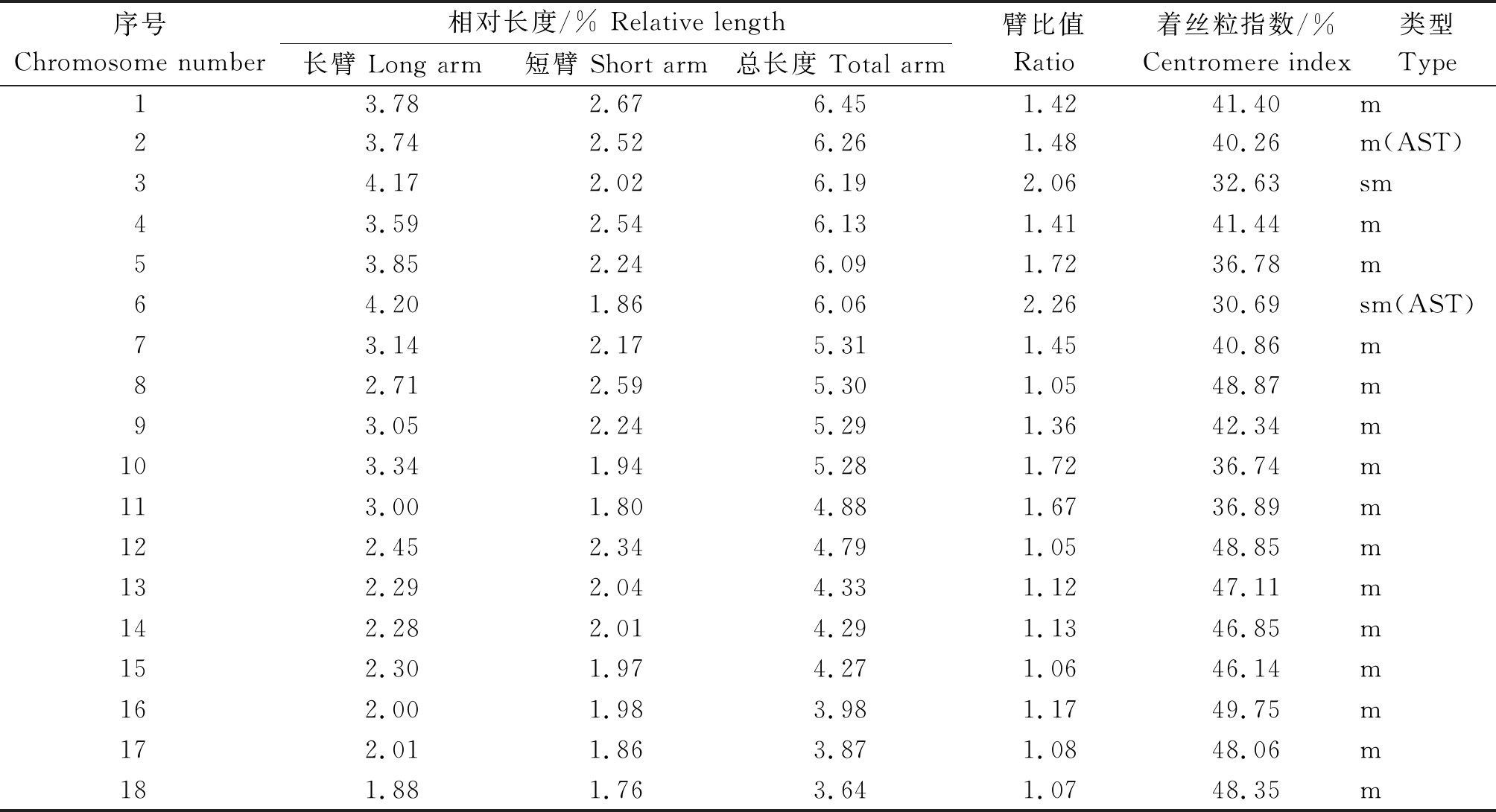
染色体计数结果表明,黑藜麦的体染色体数目为36条,染色体分成4组:L组(第1对)、M2组(第2,3,4,5,6,7,8,9,10对)、M1组(第11,12,13,14,15,16对)、S组(第17,18对);其染色体相对长度组成为4n=36=2L+18M2+12M1+4S。染色体核型公式为:2n=36=32m(4AST)+4sm,由32条中部着丝粒染色体,4条近中部着丝粒染色体组成;染色体总长度的相对长度变化为3.64%~6.45%,着丝粒指数为30.69%~49.75%,臂比值为1.05~2.26,最长染色体与最短染色体之比为2.39;臂比大于2的染色体有4条,占全染色体的11.11%,核型分类属于2B类型,核型不对称系数为58.20%。另外,还观察到在2,6号染色体的短臂上具有随体(表1、图2)。
表1 黑藜麦的染色体参数
Tab.1 Chromosome parameters of black quinoa
注:随体长度未计算在内。表2-3同。
Note:The length of the satellites were not taken into account.The same as Tab.2-3.
序号 Chromosome number 相对长度/% Relative length长臂 Long arm短臂 Short arm总长度 Total arm臂比值Ratio着丝粒指数/%Centromere index 类型Type13.782.676.451.4241.40m23.742.526.261.4840.26m(AST)34.172.026.192.0632.63sm43.592.546.131.4141.44m53.852.246.091.7236.78m64.201.866.062.2630.69sm(AST)73.142.175.311.4540.86m82.712.595.301.0548.87m93.052.245.291.3642.34m103.341.945.281.7236.74m113.001.804.881.6736.89m122.452.344.791.0548.85m132.292.044.331.1247.11m142.282.014.291.1346.85m152.301.974.271.0646.14m162.001.983.981.1749.75m172.011.863.871.0848.06m181.881.763.641.0748.35m
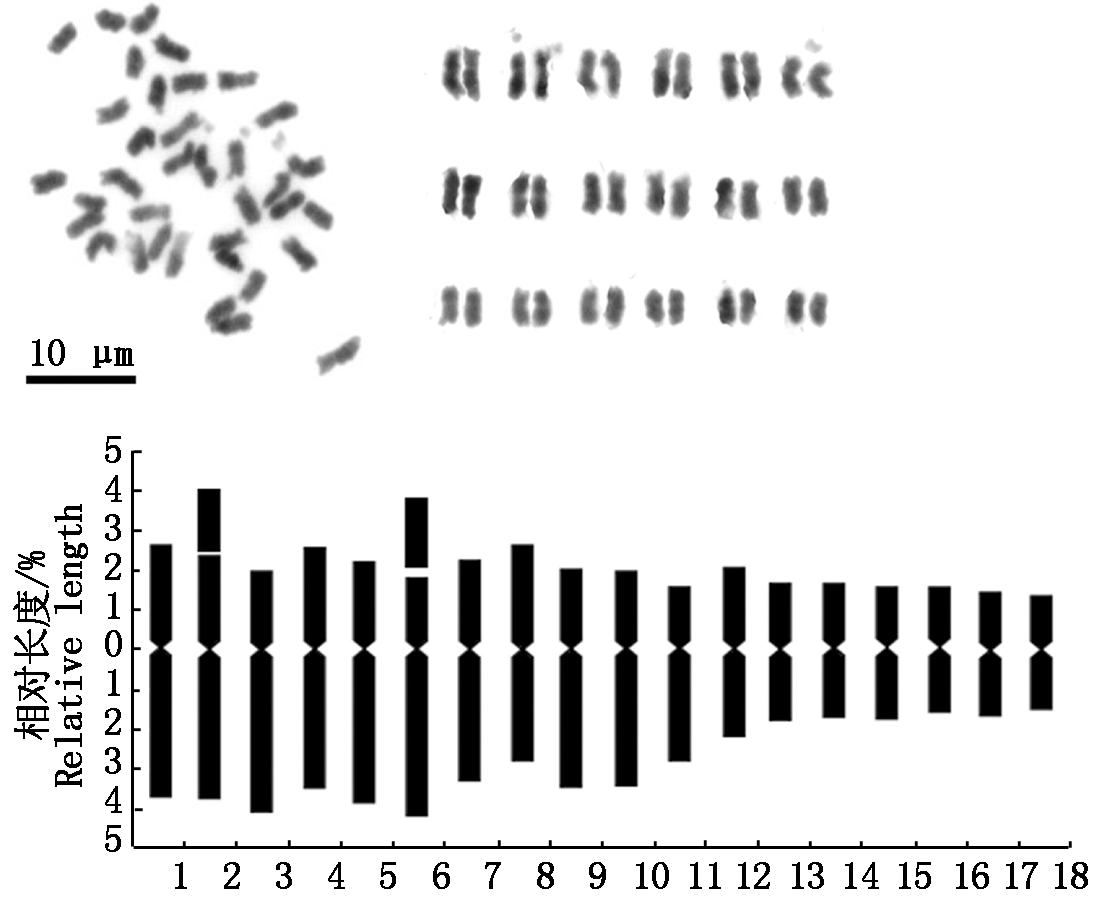
图2 黑藜麦的核型图及核型模式图
Fig.2 Karyotype and idiogram of black quinoa
2.2 白藜麦的染色体核型
体细胞染色体数目为36条。其染色体分成3组:L组(第1对)、M2组(第2,3,4,5,6,7,8,9,10对)、M1组(第11,12,13,14,15,16,17,18对);染色体相对长度组成为4n=36=2L+20M2+14M1。染色体核型公式为:2n=36=34m(4AST)+2sm,由34条中部着丝粒染色体,2条近中部着丝粒染色体组成;染色体总长度的相对长度变化为4.57%~7.88%,着丝粒指数为31.35%~48.96%,臂比值为1.04~2.19,最长染色体与最短染色体之比为2.44;臂比大于2的染色体有2条,占全染色体的5.56%,核型分类属于2B类型,核型不对称系数为58.22%。在3,5号染色体的短臂上观察到随体(表2、图3)。
表2 白藜麦的染色体参数
Tab.2 Chromosome parameters of white quinoa
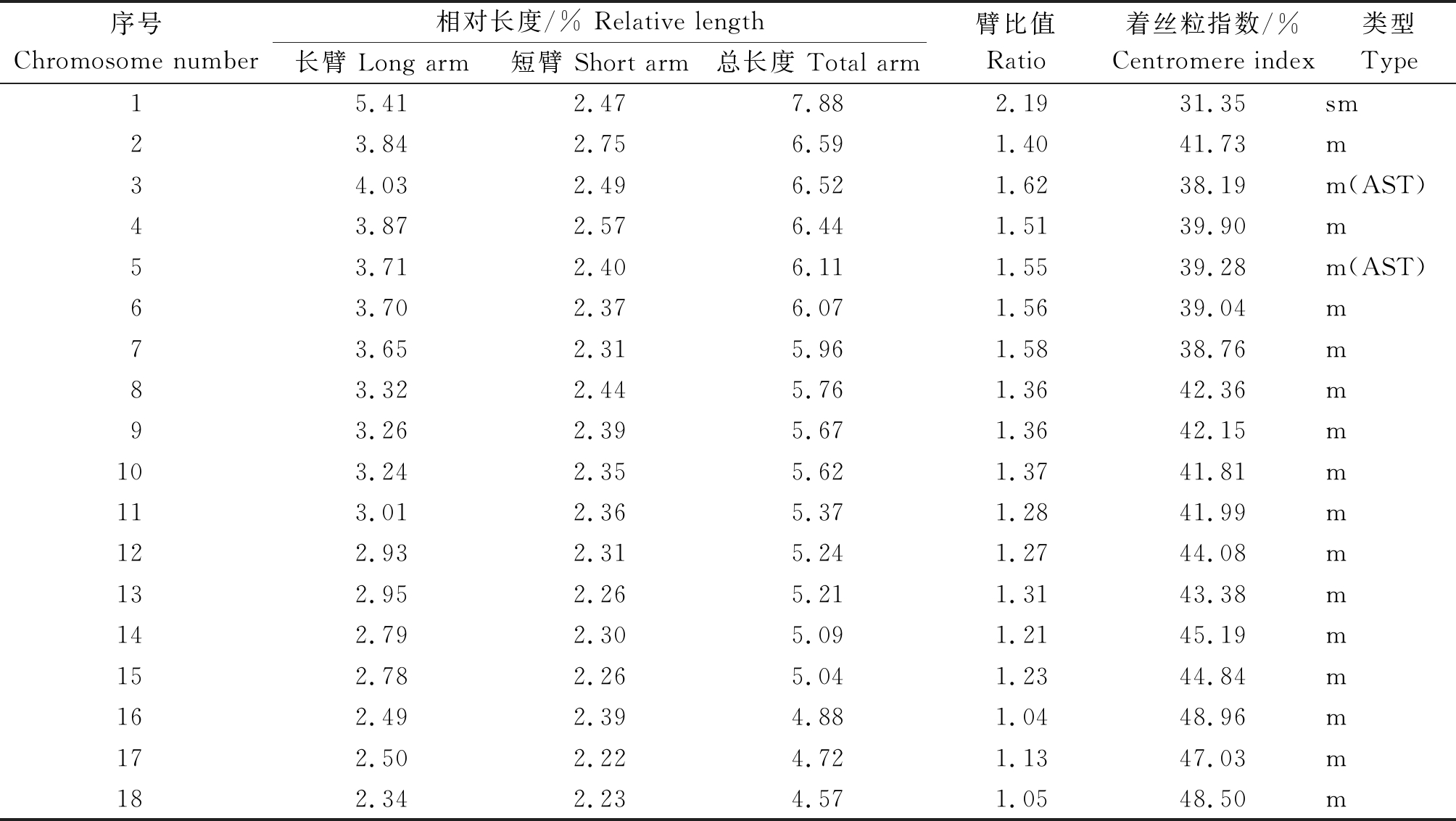
序号Chromosome number 相对长度/% Relative length长臂 Long arm短臂 Short arm总长度 Total arm 臂比值Ratio着丝粒指数/%Centromere index类型Type15.412.477.882.1931.35sm23.842.756.591.4041.73m34.032.496.521.6238.19m(AST)43.872.576.441.5139.90m53.712.406.111.5539.28m(AST)63.702.376.071.5639.04m73.652.315.961.5838.76m83.322.445.761.3642.36m93.262.395.671.3642.15m103.242.355.621.3741.81m113.012.365.371.2841.99m122.932.315.241.2744.08m132.952.265.211.3143.38m142.792.305.091.2145.19m152.782.265.041.2344.84m162.492.394.881.0448.96m172.502.224.721.1347.03m182.342.234.571.0548.50m
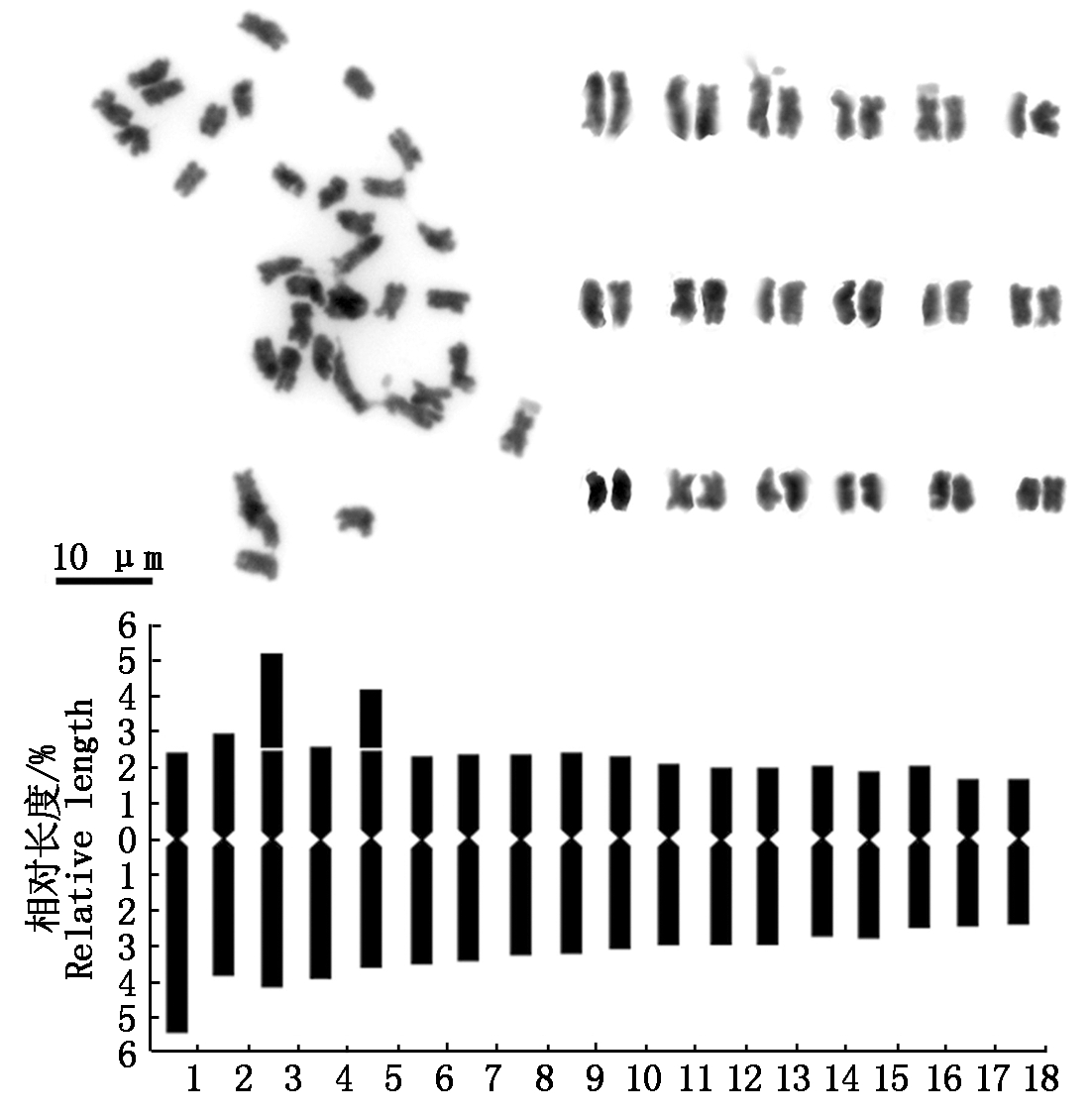
图3 白藜麦的核型图及核型模式图
Fig.3 Karyotype and idiogram of white quinoa
2.3 红藜麦的染色体核型
体细胞染色体数目也为36条。其染色分成4组:L组(第1,2,3对)、M2组(第4,5,6,7对)、M1组(第8,9,10,11,12,13,14对)、S组(第15,16,17,18对);其染色体相对长度组成为4n=36=6L+8M2+14M1+8S。藜麦染色体核型公式为:2n=36=34m(4AST)+2sm,由34条中部着丝粒染色体,2条近中部着丝粒染色体组成,染色体总长度的相对长度变化为2.28%~5.89%,着丝粒指数为28.40%~50.92%,臂比值为1.04~2.52,最长染色体与最短染色体之比为3.26;臂比大于2的染色体有2条,占全染色体的5.56%,核型分类属于2B类型,核型不对称系数为58.72%。在2,8号染色体的其中一条同源染色体的短臂上观察到随体(表3、图4)。
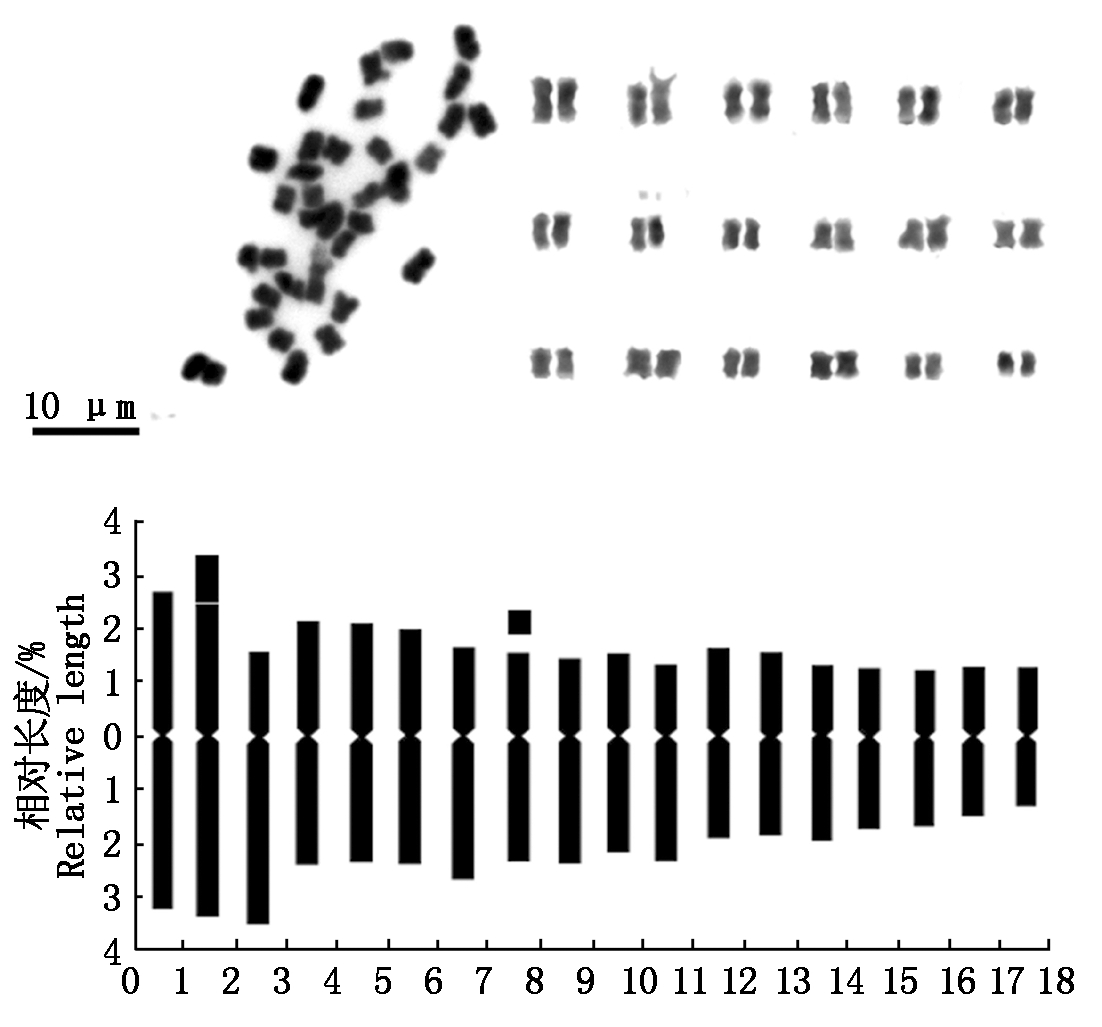
图4 红藜麦的核型图及核型模式图
Fig.4 Karyotype and idiogram of red quinoa
3 讨论
藜麦中期染色体的制片方法不尽相同,尤其是预处理的方法差别较大,例如,Bhargava等[19]用8-羟基喹啉水溶液预处理,结果较好;何燕等[20]采用0.002 mol/L 8-羟基喹啉、0.05%秋水仙素、冰水混合物和0.1%秋水仙素等预处理,发现用0.1%秋水仙素在5 ℃条件下离体根尖处理3 h效果最佳,本试验中试过将藜麦用笑气(NO)在0.5~0.8 MPa下处理2.0,2.5,3.0 h等,但结果不理想。而在4 ℃冰水混合物下处理24 h,取出后放置于卡诺固定液(无水乙醇∶冰乙酸=3∶1)中至少30 min,就可以在45%醋酸溶液下制片,效果好,相比于前两者制备中期染色体的步骤,此方法更安全、省时,简便、省试剂。
表3 红藜麦的染色体参数
Tab.3 Chromosome parameters of red quinoa
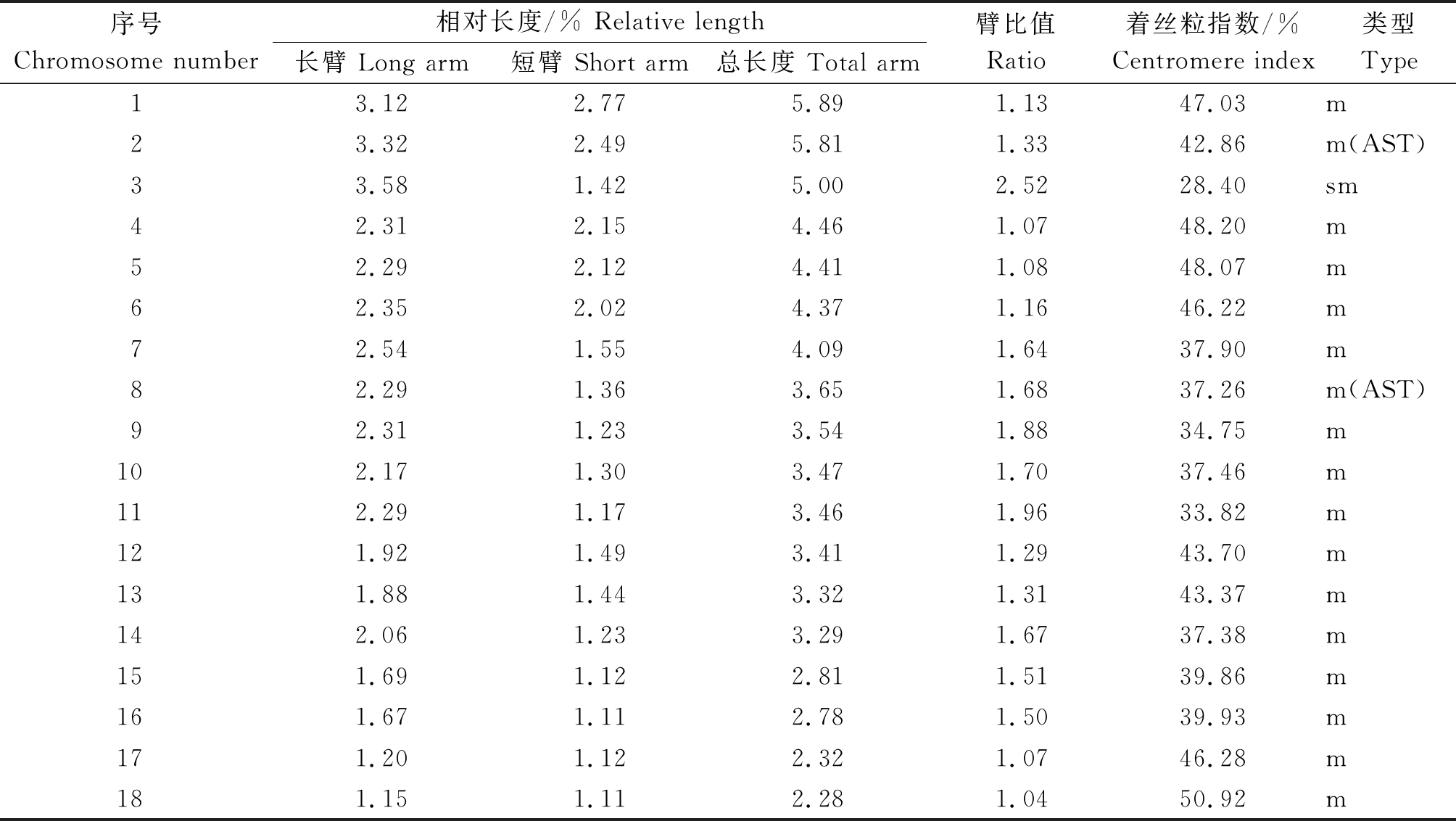
序号Chromosome number 相对长度/% Relative length长臂 Long arm短臂 Short arm总长度 Total arm臂比值Ratio着丝粒指数/%Centromere index类型Type13.122.775.891.1347.03m23.322.495.811.3342.86m(AST)33.581.425.002.5228.40sm42.312.154.461.0748.20m52.292.124.411.0848.07m62.352.024.371.1646.22m72.541.554.091.6437.90m82.291.363.651.6837.26m(AST)92.311.233.541.8834.75m102.171.303.471.7037.46m112.291.173.461.9633.82m121.921.493.411.2943.70m13 1.881.443.321.3143.37m142.061.233.291.6737.38m151.691.122.811.5139.86m161.671.112.781.5039.93m171.201.122.321.0746.28m181.151.112.281.0450.92m
本试验中3个不同籽粒颜色藜麦品种的染色体数和Bhargava等[19]及何燕等[20]的研究结果一致,均为36条,均由中部着丝粒染色体和近中部着丝粒染色体组成,染色体基数为9,均为四倍体,未发现非整倍体或多倍体现象。植物进化过程中,核型趋向于向不对称的方向发展[21]。本研究中3个品种的核型不对称系数相似,分别为58.20%,58.22%,58.72%,并且都属于2B类型,说明这些藜麦品种较为进化。3个品种中均含有2对随体,其中,黑藜麦中随体分别在2,6号染色体上,而白藜麦中随体位于3,5号染色体上,红藜麦中则在2,8号上,而目前国内外关于藜麦核型研究报道中大部分情况下随体都位于1对染色体上,大多位于2,3,8,12,15号的短臂上,也有品种中出现2对随体的情况。这种品种间的差异可能与藜麦异源起源[22]、自花授粉[23]导致染色体形态变化(主要是倒位和易位)、外部环境变化有关。
[1] Christian E Z,Vasco C,Jorge P,Luz G P,Bettit S R,Jose A T,Martha I,Kristian H L,Ursula G B.Estimation of composition of quinoa(Chenopodium quinoa Willd.)grains by near-infrared transmission spectroscopy[J].LWT-Food Science and Technology,2017,79 :126-134.doi: 10.1016/j.lwt.2017.01.026.
[2] Friedman M.Nutritional value of proteins from different food sources.a review[J].Journal of Agriculture and Food Chemistry,1996,44(1):6-29.doi:10.1021/jf9400167.
[3] Alvarez-Jubete L, Arendt E K, Gallagher E. Nutritive value and chemical composition of pseudocereals as gluten-free ingredients[J].International Journal of Food Sciences and Nutrition,2009,60(4):240-257.doi: 10.1080/09637480902950597.
[4] Stefano C, Antonella B, Lucia B, Mirella Z, Carlo V L C, Graziella A.The content of proteic and non-proteic(free and protein-bound)tryptophan in quinoa and cereal flours [J].Food Chemistry,2007,100(4):1350-1355.doi: 10.1016/j.foodchem.2005.10.072.
[5] Thoufeek A, inghal S, Kulkarni R, Pal M. Physicochemical and functional properties of Chenopodium quinoa starch[J].Carbothydrate Polymers,1996,31(1):99-103.doi: 10.1016/S0144-8617(96)00034-3.
[6] Miranda M,Vega-Galvez A,Jorquera E,Lopez J,Martinez E A.Antioxidant and antimicrobial activity of quinoa seeds(Chenopodium quinoa Willd.)from three geographical zones of Chile[M].Boca Raton:Brown Walker Press,2013:83-86.
[7] Hitomi A, Chen Y C,Tang H J, Mayumi S, Katsumi W, Toshio M. Food components in fractions of quinoa seed [J].Food Science and Technology,2002,8(1):80-84.doi: 10.3136/fstr.8.80.
[8] González J, Bruno M, Valoy M, Prado F.Genotypic variation of gas exchange parameters and leaf stable carbon and nitrogen isotopes in ten quinoa cultivars grown under drought[J].Journal of Agronomy and Crop Science,2011,197(2):144-151.doi:10.1111/j.1439-037X.2010.00446.x.
[9] González J, Bruno M, Valoy M, Prado F.Quinoa-a review[J].Czech Journal of Food Science,2009,27(27):71-79.doi:http://dx.doi.org/.
[10] 任贵兴,译.藜麦研究进展和可持续生产[M].北京:科学出版社,2018:6-100.
Translated by Ren G X. Quinoa improvement and sustainable production[M].Beijing:Science Publishing House,2018:6-100.
[11] 刘文瑜,何斌,杨发荣,吕玮,王旺田,黄杰,魏玉明,金茜,陈玉祥.不同品种藜麦幼苗对干旱胁迫和复水的生理响应[J].草业科学,2019,36(10):2656-2666.doi:10.11829/j.issn.1001-0629.2018-0698.
Liu W Y,He B,Yang F R,Lü W,Wang W T,Huang J,Wei Y M,Jin Q,Chen Y X.Physiological responses of different varieties of Chenopodium quinoa seedlings to drought stress and rehydration[J].Pratacultural Science,2019,36(10):2656-2666.
[12] 杨发荣,刘文瑜,黄杰,魏玉明.河西地区2个藜麦品种引种试验研究[J].草地学报,2018,26(5):1273-1276.doi:10.11733/j.issn.10070435.2018.05.034.
Yang F R,Liu W Y,Huang J,Wei M Y.Experimental study on introduction of two quinoa varieties in Hexi region[J]. Acta Grassland Sinica,2018,26(5):1273-1276.
[13] 魏玉明,杨发荣,刘文瑜,黄杰,金茜,王昶.陇东旱塬区复种不同藜麦品种(系)的适应性初步评价[J].西北农业学报,2020,29(5):674-686.doi:10.7606/j.issn.1004-1389.2020.05.004.
Wei Y M,Yang F R,Liu W Y,Huang J,Jin Q,Wang C.Preliminary evaluation on the adaptability of multiple cropping different quinoa varieties(Lines)in the arid Plateau of Eastern Gansu[J].Acta Agriculturae Boreali-occidentalis Sinica,2020,29(5):675-686.
[14] 陆敏佳,蒋玉蓉,陆国权,陈国林,毛前.利用SSR标记分析藜麦品种的遗传多样性[J].核农学报,2015,29(2):260-269.doi:1000-8551(2015)02-0260-10.
Lu M J,Jiang Y R,Lu G Q,Chen G L,Mao Q.Analysis of genetic diversity of Chenopodium by SSR Marker[J].Journal of Nuclear Agricultural Sciences,2015,29(2):260-269.
[15] 喻凤.苜蓿近缘种属的比较细胞遗传学研究[D].北京:中国科学院大学,2017.
Yu F.Comparative cvtogentics on Medicago sttiva L.and related species[D].Beijing:University of Chinese Academy of Sciences,2017.
[16] 李懋学,陈瑞阳.关于植物核型分析的标准化问题 [J].植物科学学报,1985,3(4):297-302.
Li M X,Chen R Y.A suggestion on the standarization of karyotype analysis in plant[J].Journal of Plant Science, 1985,3(4):297-302.
[17] Levan A,Fredga K,Sagndberg A.Nomenclature for centromeric position on chromosomes[J].Hereditas, 2009,52(2):201-220.doi:10.1111/j.1601-5223.1964.tb01953.x.
[18] Arano H.Cytological studies in subfamily carduoideae( compositae)of Japan Ⅶ[J].Shokubutsugaku Zasshi,1962,75(892):401-410.doi:10.15281/jplantres1887.76.219.
[19] Bhargava A,Shukla S,Ohri D.Karyotypic studies on some cultivated and wild species of Chenopodium(Chenopodiaceae)[J].Genetic Resources and Crop Evolution, 2006,53(7):1309-1320.doi:10.1007/s10722-005-3879-8.
[20] 何燕,邓永辉,李梦寒,冯西博,卓嘎.藜麦品系的染色.数目及核型分析[J].西南大学学报(自然科学版),2019,41(1):27-31.doi:10.13718/j.cnki.xdzk.2019.01.004.
He Y,Deng Y X,Li M H,Feng X B,Zhuo G.Chromosome number and karyotype analysis of quinoa(Chenopodium quinoa Willd.)[J].Journal of Southwest University(Natural Science Edition),2019,41(1):27-31.
[21] 魏尊征,殷选红,熊敏,王贤,周涤.3个彩色马蹄莲引进品种的核型分析[J].植物遗传资源学报,2012,13(4):650-654.doi:10.3969/j.issn.1672-1810.2012.04.024.
Wei Z Z,Yin X H,Xiong M,Wang X,Zhou D.Karyotypic analysis of three Zantedeschia hybrid cultivars [J].Journal of Plant Genetic Resources,2012,13(4):650-654.
[22] Wilson H D.Crop/weed gene flow:Chenopodium quinoa Willd.and C.berlandieri Moq.[J].Theoretical and Applied Genetic,1990,86:642-648.doi:10.1007/BF00838721.
[23] Risi J,Galwey N W.The Chenopodium grains of the andes:Inca crops for modern agriculture.in advances in applied biology[M].London:Academic press,1990:145-216.