气候变化和不合理的灌溉使全球盐碱地的面积日趋增加。据统计,全世界盐碱地的面积约为9.54亿hm2,且每年以1.0×106~1.5×106hm2的速度增长[1]。松嫩平原西部是我国最主要的内陆盐碱地集中区之一,同时也是世界三大片苏打盐渍土集中分布区之一,盐碱化土地面积300万hm2。盐碱地种稻是改良盐碱地最有效的手段之一,所以改良水稻耐盐、碱性是盐碱地种稻的前提。已有研究表明,水稻苗期对盐、碱胁迫最为敏感[2],而苗期迅速返青成活是盐碱地种稻的关键。盐胁迫主要造成离子毒害和渗透胁迫,增加土壤渗透势[3],使植株体内的Na+含量升高,并影响K+的吸收,引起离子平衡失调,进而引起植物体内的各种生理变化[4]。碱胁迫会使植株变矮,叶面积指数变小,分蘖减少,叶绿素含量降低,进而生物学产量和经济产量下降[5-7]。盐、碱双重胁迫则会阻碍水稻的生长发育,使细胞代谢减慢,光合作用和呼吸作用受阻。过度盐碱害甚至还会导致水稻死亡[8]。综上,盐、碱胁迫不仅会限制水稻的生产面积进一步扩大,而且还会降低水稻产量。
近年来,国内外研究学者对水稻的耐盐、碱性进行了大量的QTL定位研究。Sabouri等[9]定位到与耐盐等级、苗高、和苗干质量相关的6个QTL。Thomson等[10]利用NILs群体定位到了27个控制水稻苗期耐盐相关性状的QTL。祁栋灵[11]利用碱胁迫高产106/长白9号的F2:3家系群体,检测到7个影响水稻发芽率的QTL,6个影响水稻碱害率的QTL。杨静[12]利用Lemont和特青的双向回交导入系群体,检测到了18个与耐盐性相关的QTL。Lee等[13]利用NILs群体定位到2个影响水稻苗期盐害级别的QTL。顾兴友等[14]利用Pokkali和Peta的BC1群体,检测到4个影响水稻苗期和成熟期耐盐性的QTL。邢军等[15]利用东农425和长白10号的RIL群体,检测到5个耐盐QTL和10个耐碱QTL。目前,在盐胁迫下对水稻Na+、K+浓度的QTL定位研究较多,但在碱胁迫下的相关研究少有报道,同一试验中在盐、碱胁迫条件下同时进行研究和比较的分析更是鲜见报道。因此,在盐、碱胁迫条件下同时对水稻Na+、K+的遗传机制进行研究具有重大意义,更为耐盐、碱水稻的品种选育奠定基础。
本研究利用小白粳子/空育131衍生的含有200个株系的RIL群体,及其包含142个SSR标记的遗传连锁图谱对盐、碱胁迫条件下水稻苗期盐、碱害级别、地上部Na+浓度、K+浓度及Na+/K+进行QTL分析,旨在进一步获得耐盐、碱主效QTL,为水稻耐盐、碱性QTL的精细定位和分子辅助选择育种提供科学依据并奠定理论基础。
1 材料和方法
1.1 试验材料
以陆稻品种小白粳子为母本,耐盐、碱水稻品种空育131为父本配置杂交组合,通过“单粒传”得到了包含200个家系的F9群体材料。
1.2 水稻苗期耐盐性和耐碱性鉴定
试验于东北农业大学农学院水稻育种研究室内进行,试验共设盐胁迫和碱胁迫2种处理。选取籽粒饱满的种子在50 ℃烘干48 h打破休眠,用0.5%次氯酸钠处理10 min进行表面消毒,再用清水冲洗3次后催芽。每个家系及亲本种子选取24粒露白发芽一致种子播种于底部剪孔的PCR板中,置于15 L盛有水的塑料盘中,用pH值5.5左右的清水培养至三叶一心期。盐胁迫将清水换成含120 mmol/L NaCl的营养液处理12 d,营养液的pH维持在5.5左右,碱胁迫将清水换成40 mmol/L NaHCO3营养液处理12 d,pH维持在8.5左右,每2 d换1次营养液。在盐、碱胁迫12 d后,每个株系选取10株测量盐、碱害级别,盐、碱害级别规定为1~9级(具体分级标准见表1)。然后收获每个株系的地上部分,105 ℃杀青,随后在80 ℃条件下烘干至恒质量,每个品种称取0.5 g剪碎的烘干苗放入试管中,然后加入10 mL浓度为1 mmol/L的稀盐酸溶液,于55 ℃振荡培养箱(ZHSY-50N型,由上海知楚仪器有限公司制造)中振荡提取3 h,将样品稀释后用火焰光度计(M410型,由上海仪电分析仪器有限公司制造)分别测定Na+、K+浓度并进一步计算Na+/K+(性状缩写见表2)。
表1 水稻苗期盐、碱害级别的评价标准
Tab.1 Evaluation criteria for score of salt and alkali toxicity at seedling stage of rice

盐碱害级别Score of salt and alkali toxicity考察标准Visual symptoms耐性评价Tolerance1幼苗生长正常,叶片没有受害症状高抗3幼苗几乎正常生长,但是叶尖和少数叶片发白、卷曲抗5幼苗生长轻度受阻,小部分叶片卷曲中抗7秧苗生长中度受阻,少数叶片干枯感9秧苗完全停止发生,大部分植株接近死亡高感
表2 性状全称及缩写
Tab.2 Characters full name and abbreviation
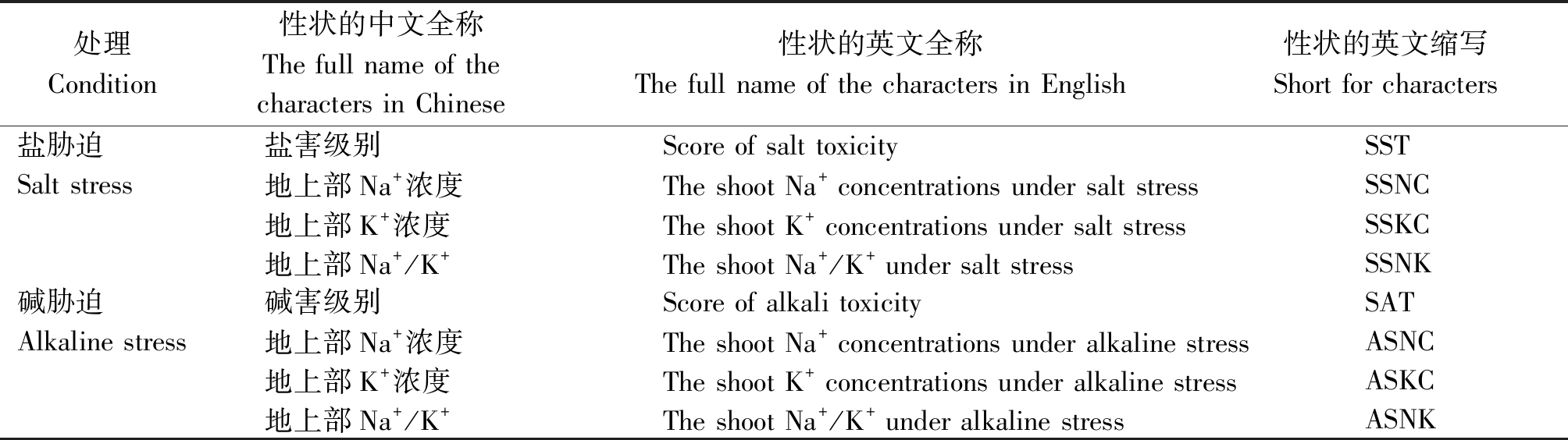
处理Condition性状的中文全称The full name of the characters in Chinese性状的英文全称The full name of the characters in English性状的英文缩写Short for characters盐胁迫盐害级别Score of salt toxicity SSTSalt stress地上部Na+浓度The shoot Na+ concentrations under salt stressSSNC地上部K+浓度The shoot K+ concentrations under salt stressSSKC地上部Na+/K+The shoot Na+/K+ under salt stressSSNK碱胁迫碱害级别Score of alkali toxicity SATAlkaline stress地上部Na+浓度The shoot Na+ concentrations under alkaline stressASNC地上部K+浓度The shoot K+ concentrations under alkaline stressASKC地上部Na+/K+The shoot Na+/K+ under alkaline stressASNK
1.3 遗传连锁图谱的构建及 QTL 分析
本研究在Xing等[16]利用两亲本衍生的F6 RIL群体构建的包含104个SSR分子标记的遗传图谱基础上新增38个SSR标记。利用Mapmaker 3.0软件构建图谱,使用Kosambi函数将重组率转化为遗传距离(cM),新图谱全长1 290.25 cM,平均标记间距为9.09 cM。采用 Mapchart 2.2 进行遗传连锁图谱的绘制,利用 IciMapping v3.3的完备区间作图法(ICIM)进行QTL定位[17],LOD=2.5为QTL的阈值,QTL的命名原则遵循McCouch等[18]方法。
2 结果与分析
2.1 水稻盐、碱胁迫条件下苗期 Na+、K+浓度分析
通过对亲本及重组自交系群体在盐、碱胁迫条件下盐、碱害级别、地上部Na+、K+浓度及 Na+/K+的统计分析,可得出以下结果(表3)。在盐、碱胁迫条件下SST、SSNC、SSKC、SSNK、SAT、ASNC、ASKC、ASNK在空育131和小白粳子两亲本间均呈极显著差异。RIL群体中SST高于SAT,表明RIL群体相对盐胁迫更耐碱胁迫;SSNC和SSKC分别高于ASNC和ASKC,但ASNK高于SSNK;SSNC数据变异范围最大,为7.83~35.02,其次为ASNC,变异范围为5.87~20.01,SSNK和ASNK的变异系数最大,分别为37.65%和32.47%;除ASKC外,重组自交系的其他性状的平均值均介于小白粳子和空育131之间且存在明显的超亲分离现象。对数据进行正态分布的适合性检验,发现数据基本都符合正态分布,表现出了典型的数量性状遗传特征,符合QTL定位的要求(图1)。
表3 盐、碱胁迫条件下水稻亲本及重组自交系群体的表型性状
Tab.3 Phenotypic characters of rice parents and recombinant inbred populations under salt and alkaline stress
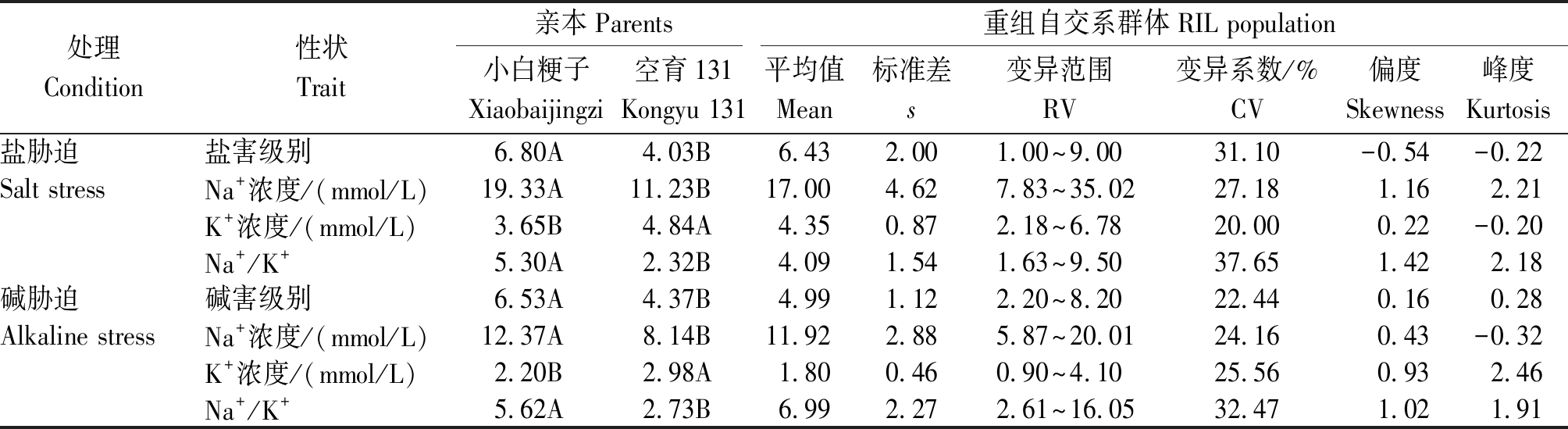
处理Condition性状Trait亲本Parents重组自交系群体RIL population小白粳子Xiaobaijingzi空育131Kongyu 131平均值Mean标准差s变异范围RV 变异系数/%CV偏度Skewness峰度Kurtosis盐胁迫盐害级别6.80A4.03B6.432.001.00~9.0031.10-0.54-0.22Salt stressNa+浓度/(mmol/L)19.33A11.23B17.004.627.83~35.0227.181.162.21K+浓度/(mmol/L)3.65B4.84A4.350.872.18~6.7820.000.22-0.20Na+/K+5.30A2.32B4.091.541.63~9.5037.651.422.18碱胁迫碱害级别6.53A4.37B4.991.122.20~8.2022.440.160.28Alkaline stressNa+浓度/(mmol/L)12.37A8.14B11.922.885.87~20.0124.160.43-0.32K+浓度/(mmol/L)2.20B2.98A1.800.460.90~4.1025.560.932.46Na+/K+5.62A2.73B6.992.272.61~16.0532.471.021.91
注:不同大写字母表示在0.01水平上差异极显著。
Note:Different capital letter indicate extremely significant difference at the level of 0.01.
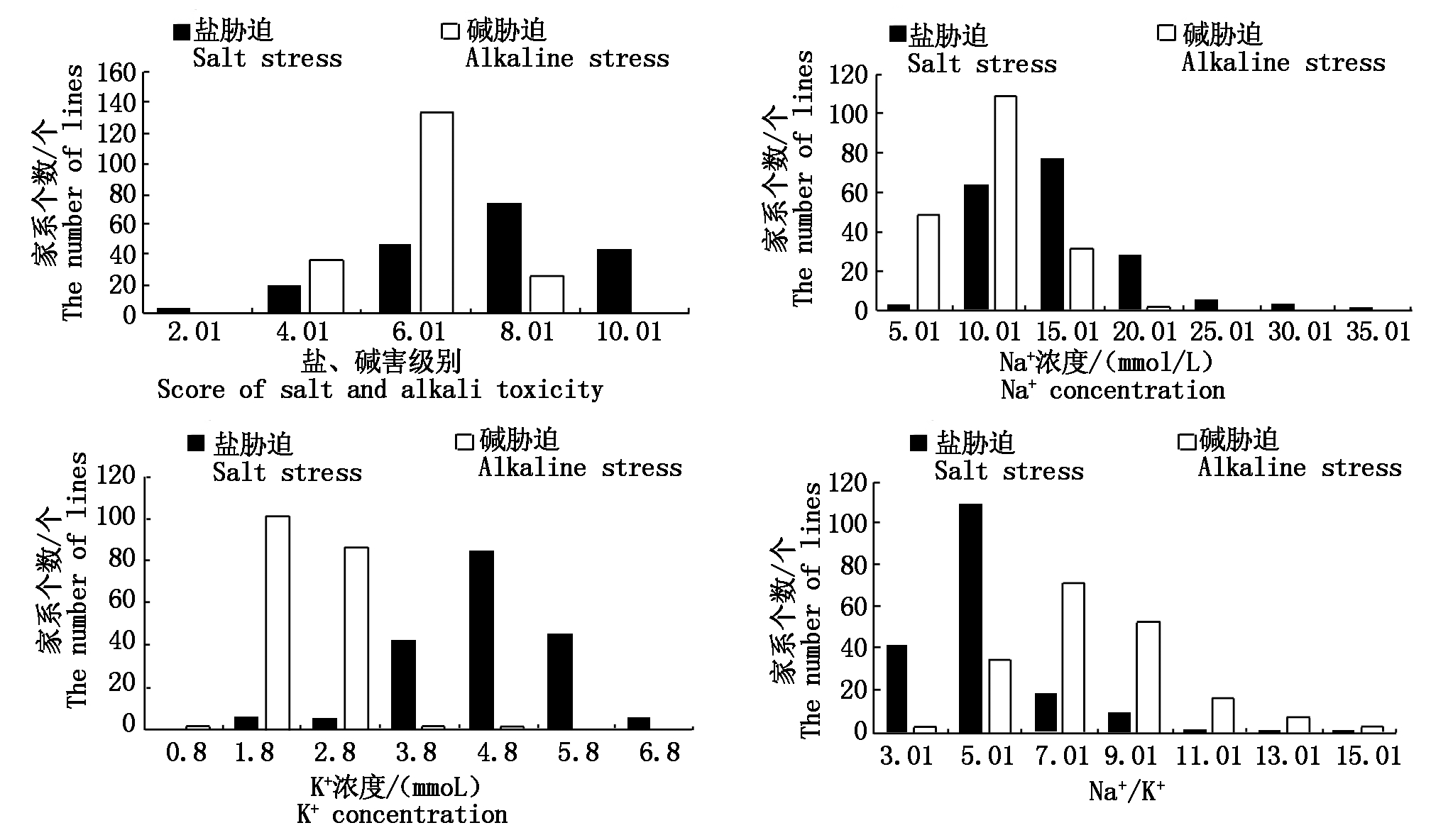
图1 盐、碱胁迫下盐碱害级别、地上部Na+浓度、K+浓度、Na+/K+的分布
Fig.1 The distribution of score of salt toxicity and alkali toxicity,the shoot Na+ concentrations, K+ concentrations,Na+/K+under salt stress and alkaline stress
2.2 水稻盐、碱胁迫条件下 Na+、K+相关表型性状的相关性分析
对盐、碱胁迫条件下SST、SSNC、SSKC、SSNK、SAT、ASNC、ASKC、ASNK等8个性状进行相关分析(表4)。盐胁迫下,SSNK与SSNC呈极显著正相关,而SSNK与SSKC呈极显著负相关,SST与SSNC、SSNK呈显著正相关,说明地上部Na+浓度越高,盐害级别越大;碱胁迫下,ASNC与ASNK呈极显著正相关,与SAT呈显著正相关,ASKC与ASNC呈显著正相关,ASKC与ASNK呈显著负相关。SSNC与SSNK、ASNC与ASNK的相关系数较大,分别为0.821和0.629;SSNK与ASKC呈极显著正相关,相关系数为0.429。
表4 各性状间的相关系数
Tab.4 Correlation coefficients of each trait
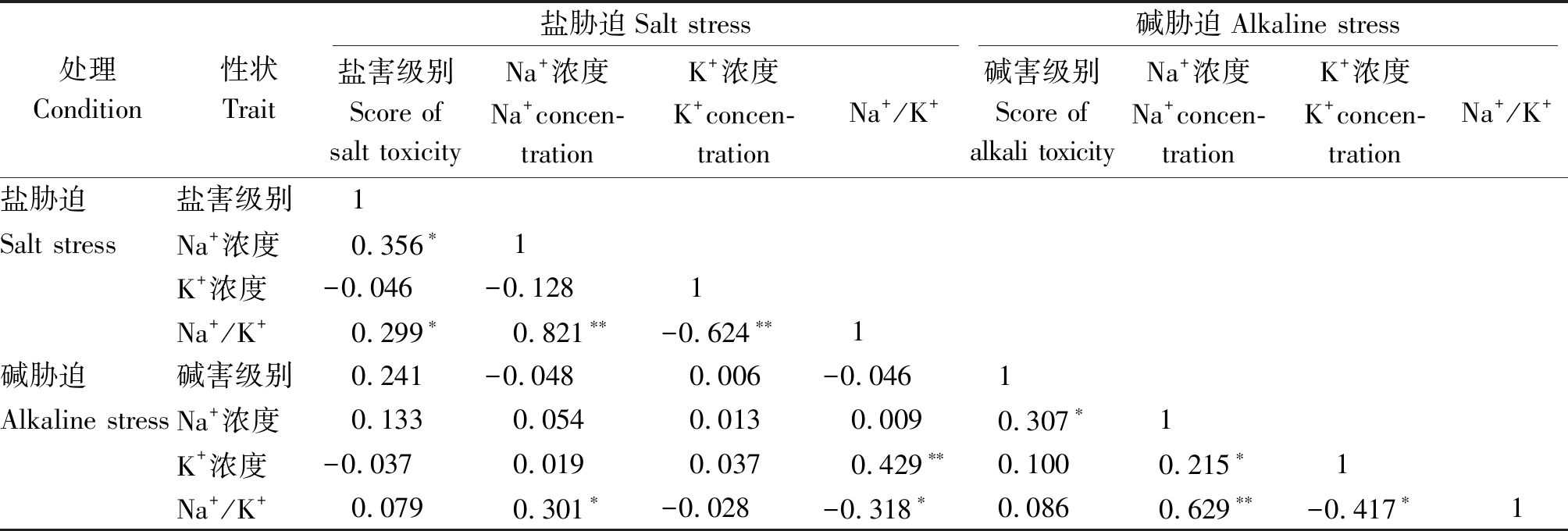
处理Condition性状Trait盐胁迫Salt stress碱胁迫Alkaline stress盐害级别Score ofsalt toxicityNa+浓度Na+concen-trationK+浓度K+concen-trationNa+/K+碱害级别Score ofalkali toxicityNa+浓度Na+concen-trationK+浓度K+concen-trationNa+/K+盐胁迫盐害级别1Salt stressNa+浓度0.356∗1K+浓度-0.046-0.1281Na+/K+0.299∗0.821∗∗-0.624∗∗1碱胁迫碱害级别0.241-0.0480.006-0.0461Alkaline stressNa+浓度0.1330.0540.0130.0090.307∗1K+浓度-0.0370.0190.0370.429∗∗0.1000.215∗1Na+/K+0.0790.301∗-0.028-0.318∗0.0860.629∗∗-0.417∗1
注:*.在0.05水平上显著相关;**.在0.01水平上极显著相关。
Note: *.Significant correlation at the level of 0.05; **.Extremely significant correlation at the level of 0.01.
2.3 水稻耐盐、碱相关性状的QTL分析
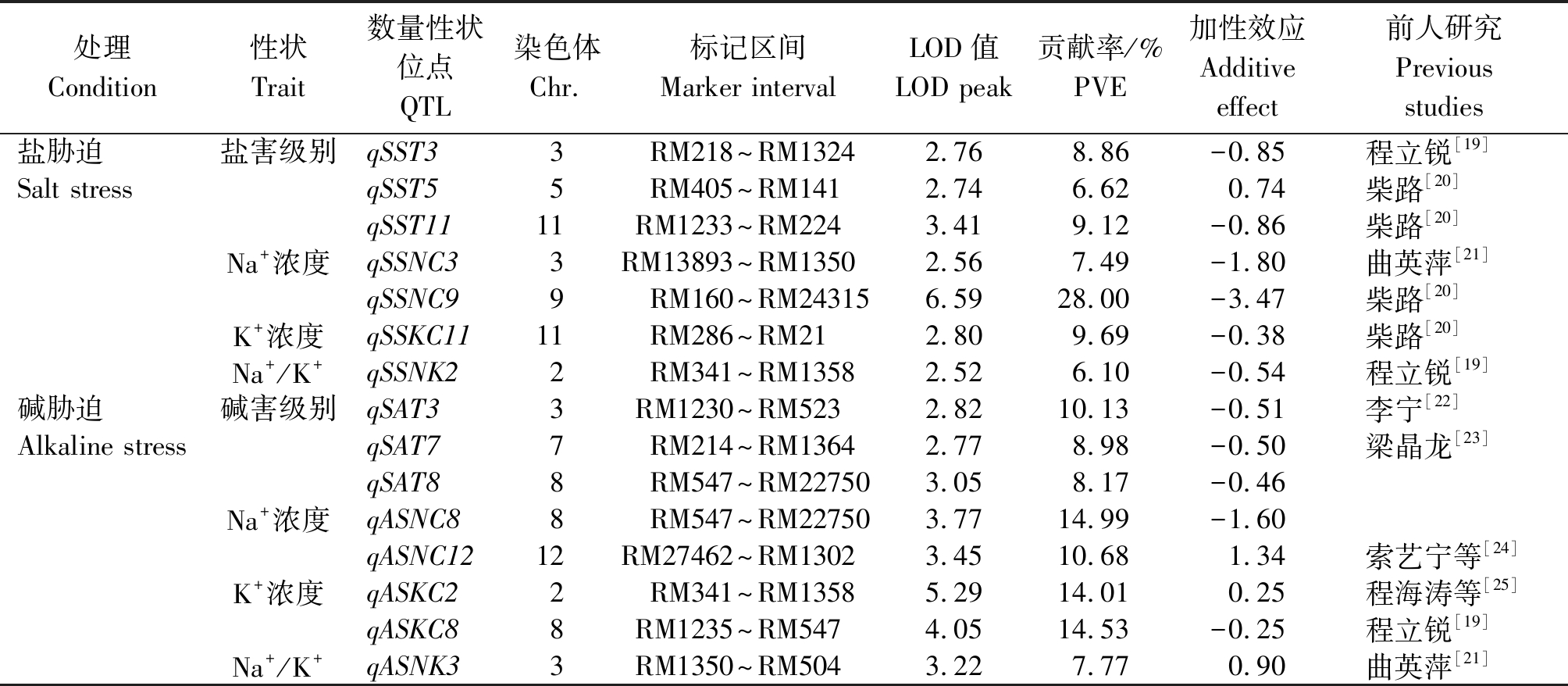
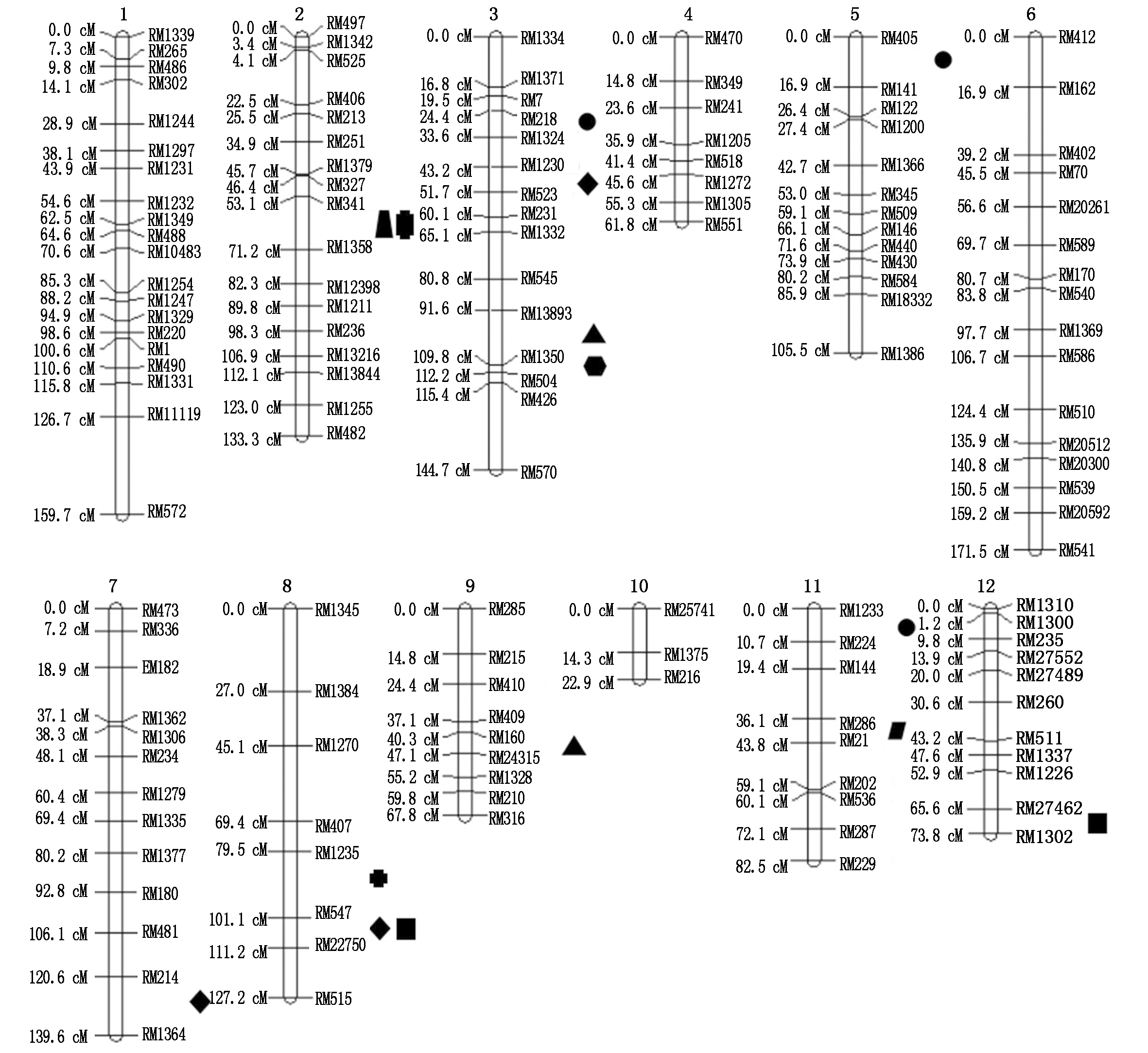
在盐、碱胁迫条件下,对SST、SSNC、SSKC、SSNK、SAT、ASNC、ASKC、ASNK等8个性状进行QTL分析(表5),2种条件下共检测到15个QTL,分布在第2,3,5,7,8,9,11,12染色体上(图2),发现有13个QTL为已知QTL,LOD值2.52~6.59,表型贡献率为6.10%~28.00%。
在盐胁迫条件下共检测到7个QTL。检测到3个与SST相关的QTL,分布在第3,5,11染色体上,其中qSST11的贡献率最大,为9.12%,增效等位基因来自于空育131;在第3和第9染色体上检测到2个与SSNC相关的QTL,其中qSSNC9的贡献率较大,为28.00%,增效等位基因来自于空育131;在第11染色体上检测到1个与SSKC相关的QTL即qSSKC11,它的增效等位基因来自于空育131;在第2染色体上检测到1个与SSNK相关的QTL即qSSNK2,它的增效等位基因来自于空育131。
在碱胁迫条件下共检测到8个QTL。在第3,
表5 水稻苗期各性状的QTL及遗传效应
Tab.5 QTLs and genetic effects of rice traits at seedling stage
处理Condition性状Trait数量性状位点QTL染色体Chr.标记区间Marker intervalLOD值LOD peak贡献率/%PVE加性效应Additiveeffect前人研究Previousstudies盐胁迫盐害级别qSST33RM218~RM13242.768.86-0.85程立锐[19]Salt stressqSST55RM405~RM1412.746.620.74柴路[20]qSST1111RM1233~RM2243.419.12-0.86柴路[20]Na+浓度qSSNC33RM13893~RM13502.567.49-1.80曲英萍[21]qSSNC99RM160~RM243156.5928.00-3.47柴路[20]K+浓度qSSKC1111RM286~RM212.809.69-0.38柴路[20]Na+/K+qSSNK22RM341~RM13582.526.10-0.54程立锐[19]碱胁迫碱害级别qSAT33RM1230~RM5232.8210.13-0.51李宁[22]Alkaline stressqSAT77RM214~RM13642.778.98-0.50梁晶龙[23]qSAT88RM547~RM227503.058.17-0.46Na+浓度qASNC88RM547~RM227503.7714.99-1.60qASNC1212RM27462~RM13023.4510.681.34索艺宁等[24]K+浓度qASKC22RM341~RM13585.2914.010.25程海涛等[25]qASKC88RM1235~RM5474.0514.53-0.25程立锐[19]Na+/K+qASNK33RM1350~RM5043.227.770.90曲英萍[21]
注:QTL命名中的第一个大写字母S和A分别代表盐胁迫和碱胁迫。
Note:The first capital letter S and A represent salt stress and alkaline stress.
![]() .分别代表盐胁迫下的SST、SSNC、SSKC、SSNK的QTL;
.分别代表盐胁迫下的SST、SSNC、SSKC、SSNK的QTL;![]() .分别代表碱胁迫下的SAT、ASNC、ASKC、ASNK的QTL。
.分别代表碱胁迫下的SAT、ASNC、ASKC、ASNK的QTL。 ![]() .QTL for SST,SSNC,SSKC,SSNK under salt stress;
.QTL for SST,SSNC,SSKC,SSNK under salt stress;![]() .QTL for SAT,ASNC,ASKC,ASNK under alkaline stress.
.QTL for SAT,ASNC,ASKC,ASNK under alkaline stress.
图2 水稻苗期相关性状的耐盐、碱性的QTL定位
Fig.2 QTL mapping of salt-tolerant and alkali-tolerant traits in rice during seedling stage
7,8染色体上检测到3个与SAT有关的QTL即qSAT3、qSAT7、qSAT8,其中qSAT3的贡献率最大,为10.13%,它的增效等位基因来自于空育131;在第8,12染色体上检测到2个有关ASNC的QTL,其中qASNC8的贡献率较大,为14.99%,增效等位基因来自于空育131;在第2和第8染色体上检测到2个与ASKC相关的QTL,其中qASKC8的贡献率最大,为14.53%,增效等位基因来自于空育131;在第3染色体上检测到1个与ASNK相关的QTL即qASNK3,它的增效等位基因来自于小白粳子。
3 讨论与结论
3.1 水稻苗期耐盐、碱性及Na+、K+的关系
水稻的耐盐、碱性是极其复杂的遗传性状,涉及水稻生长发育过程中不同阶段的一系列生理生化过程。水稻发育过程中种子萌芽期和分蘖拔节期的耐盐、碱性要强于幼苗期[26],同时水稻苗期是其受盐、碱胁迫较敏感的时期,所以在苗期对水稻进行相关性状的耐盐、碱性的检测,结果会较为准确。郑少玲等[27]认为耐盐的水稻品种自身会对Na+有积累作用,水稻地下部的Na+会抑制其向地上部运输,所以地上部的Na+较少,致使品种本身的耐盐性提高。Sharma[28]对耐碱品种和敏碱品种做了比较分析,研究发现耐碱品种体内积累了较低的Na+和较高的K+,而敏碱品种恰恰相反,说明耐碱品种体内存在一种生理机制,可以在吸收的K+同时控制Na+的进入,进而具有较高的K+/Na+。本研究中发现,盐、碱胁迫条件下水稻的SST与SAT定位在不同区间,说明水稻耐盐性和耐碱性之间可能存在不同的遗传机制,而盐胁迫下与Na+浓度相关的qSSNC3和碱胁迫下与Na+/K+相关的qASNK3定位在了相邻区间,盐胁迫下与Na+/K+相关的qSSNK2和碱胁迫下与K+浓度相关的qASKC2定位在了相同区间,说明盐、碱胁迫条件下的地上部Na+、K+浓度具有一定的遗传重叠。
3.2 盐、碱胁迫下Na+、K+的相关性与QTL分析
影响相关性状的QTL常常被定位在相同或相邻的染色体区域内[29-31]。盐胁迫下,郑洪亮等[32]的研究发现,影响水稻幼苗前期相对苗高的RSH与影响相对根数的RRN呈极显著正相关,qRSH3与qRRN3定位在了第3染色体的相同区间RM1324~RM517;碱胁迫下,邹德堂等[33]的研究表明,影响水稻抽穗期叶长的Ll与影响叶面积的LAR呈极显著正相关,qALl8与qALAR8定位在了第8染色体的相同区间RM22475~Indel66。本研究中,SSNC与ASNK 呈显著正相关,SSNK与ASKC呈极显著正相关,ASKC与ASNC呈显著正相关,SAT与ASNC呈显著正相关。同时通过QTL定位发现,qSSNC3和qASNK3定位在了相邻区间RM13893~RM1350和RM1350~RM504;qSSNK2和qASKC2定位在了相同区间RM341~RM1358;qASKC8与qASNC8定位在了相邻区间RM1235~RM547和RM547~RM22750;qSAT8和qASNC8定位在了相同区间RM547~RM22750,表明这些QTL可能有部分重叠区域或存在紧密连锁,或者为一因多效。
3.3 与前人研究结果的比较分析
到目前为止,前人已对水稻耐盐、碱性QTL定位进行了大量研究[34-36]。通过相同的SSR标记和比较图谱[37-39],将本研究定位到的苗期耐盐、耐碱QTL与前人耐盐、耐碱QTL结果进行比较,发现本研究检测到的15个QTL中有13个与前人定位在相邻或相同区间。本研究中影响苗期地上部盐害级别的qSST3与程立锐[19]在盐胁迫下检测到的影响根部Na+浓度的qSRNC3-1定位在了相邻区间;qSST5与柴路[20]在盐胁迫下检测到的影响单株产量的QTgw5定位在了相邻区间;qSST11与在盐胁迫下检测到的影响千粒质量的QGyp11定位在了相邻区间[20];qSSNC9、qSSNC3、qSSKC11、qSSNK2分别与在盐胁迫下检测到的影响单株产量的QTgw9[20]、影响发芽率的相对盐害率qSRGP3-2[21]、影响单株产量的GYP[20]、影响Na+浓度的QSnc2[19]定位在了相邻区间。影响苗期地上部K+浓度的qASKC8与在碱胁迫下检测到的影响千粒质量的GW定位在了相邻区间[19];qASNK3与曲英萍[21]在碱胁迫下检测到的影响死叶率的qADLR3-2定位在了相邻区间;qASKC2、qASNC12、qSAT3、qSAT7分别与在碱胁迫下检测到的影响幼苗前期碱害率的ADS[25]、影响根数的qN-ARN3[24]、影响地上部Na+/K+浓度的qSNK3[22]、影响发芽率相对盐害率的qAGRA7-2定位在了相邻区间[23]。这些QTL能在不同遗传背景下被检测到,表明它们是稳定的QTL。另外,本研究定位在相同区间RM547~RM22750的qSAT8和qASNC8,在前人研究中未见报道,可能是新的QTL位点,有待进一步研究。
3.4 结论
本研究在盐、碱2种胁迫条件下共定位到了15个QTL,其中盐、碱胁迫条件下分别检测到7,8个QTL,分布在第2,3,5,7,8,9,11和12染色体上,表型贡献率在6.10%~28.00%,其中13个QTL与前人定位在相邻或相同区间,而位于RM547~RM22750之间的qSAT8和qASNC8为新的QTL位点。
水稻苗期耐盐性和耐碱性之间可能存在不同的遗传机制,而盐、碱胁迫条件下地上部Na+、K+浓度具有一定的遗传重叠。
[1] 王苗,齐树亭,葛美丽.盐生植物对滨海盐渍土生物改良的研究进展[J].安徽农业科学,2008,36(7):2898-2899,2954.doi:10.13989/j.cnki.0517-6611.2008.07.045.
Wang M, Qi S T, Ge M L.Research progress on the biological improvement of the coastal saline soil by halophytes[J].Journal of Anhui Agricultural Sciences,2008,36(7):2898-2899,2954.
[2] 汪斌,兰涛,吴为人.盐胁迫下水稻苗期Na+含量的QTL定位[J].中国水稻科学,2007,21(6):585-590.doi:10.16819/j.1001-7216.2007.06.005.
Wang B, Lan T, Wu W R.Mapping of QTLs for Na+ content in rice seedlings under salt stress[J].Chinese Journal of Rice Science,2007,21(6):585-590.
[3] 刘滨硕,康春莉,王鑫,包国章.羊草对盐碱胁迫的生理生化响应特征[J].农业工程学报,2014,30(23):166-173.doi:10.3969/j.issn.1002-6819.2014.23.021.
Liu B S, Kang C L, Wang X, Bao G Z.Physiological and biochemical response characteristics of Leymus chinensis to saline-alkali stress[J].Transactions of the Chinese Society of Agricultural Engineering,2014,30(23):166-173.
[4] 张新春,庄炳昌,李自超.植物耐盐性研究进展[J].玉米科学,2002,10(1):50-56. doi:10.3969/j.issn.1005-0906.2002.01.015.
Zhang X C, Zhuang B C, Li Z C.Advances in study of salt-stress tolerance in plants[J].Journal of Maize Sciences,2002,10(1):50-56.
[5] 梁正伟,杨福,王志春,陈渊.盐碱胁迫对水稻主要生育性状的影响[J].生态环境,2004,13(1):43-46.doi:10.3969/j.issn.1674-5906.2004.01.014.
Liang Z W, Yang F, Wang Z C, Chen Y.Effect of the main growth characteristics of rice under saline-alkali stress[J].Ecology and Environment,2004,13(1):43-46.
[6] 王英,张国民,李景鹏,马军韬, 王永力,张丽艳,邓凌伟.寒地粳稻耐碱研究进展及开发前景[J].作物杂志,2016(6):1-8. doi:10.16035/j.issn.1001-7283.2016.06.001.
Wang Y, Zhang G M, Li J P,Ma J T,Wang Y L,Zhang L Y,Deng L W.Advances in alkaline tolerance of japonica rice in cold zone[J].Crops,2016 (6):1-8.
[7] 王志欣. 东北粳稻耐盐碱性种质筛选及相关性状的QTL定位[D].哈尔滨:东北农业大学,2012.
Wang Z X.Screening of salt-tolerant alkaline germplasm and QTL mapping of related traits in rice in Northeast China[D]. Harbin:Northeast Agricultural University,2012.
[8] Kumar K,Kumar M,Kim S R,Ryu H,Cho Y G.Insights into genomics of salt stress response in rice[J].Rice,2013,6(1):1.doi:10.1186/1939-8433-6-27.
[9] Sabouri H,Rezai A M,Moumeni A,Kavousi A,Katouzi M,Sabouri A.QTLs mapping of physiological traits related to salt tolerance in young rice seedlings[J].Biologia Plantarum,2009,53(4):657-662.doi:10.1007/s10535-009-0119-7.
[10] Thomson M,de Ocampo M,Egdane J A,Rahman M A,Sajise A G C,Adorada D, Tumimbang E, Blumwald E, Seraj Z I, Singh R K, Gregorio G B, Ismail A M.Characterizing the Saltol quantitative trait locus for salinity tolerance in rice[J].Rice,2010,3(2):148-160.doi:10.1007/s12284-010-9053-8.
[11] 祁栋灵.水稻耐碱性数量性状座位(QTLs)初步分析[D].雅安:四川农业大学,2006.
Qi D L.Preliminary analysis of rice alkali tolerance quantitative trait loci (QTLs)[D].Yaan:Sichuan Agricultural University,2006.
[12] 杨静.利用双向导入系剖析水稻耐盐QTL定位的遗传背景效应[D]. 哈尔滨:东北农业大学,2009.doi:10.7666/d.d072699.
Yang J.Analysis of genetic background effects of salt-tolerant QTL mapping in rice using two-way introduction[D]. Harbin:Northeast Agricultural University,2009.
[13] Lee S Y, Ahn J H, Cha Y S, Yun D W, Lee M C, Ko J C, Lee K S,Eun M Y. Mapping QTLs related to salinity tolerance of rice at the young seedling stage[J].Plant Breeding,2007,126(1): 43-46. doi:10.1111/j.1439-0523.2007.01265.x.
[14] 顾兴友,梅曼彤,严小龙,郑少玲,卢永根.水稻耐盐性数量性状位点的初步检测[J].中国水稻科学,2000,14(2):65-70. doi:10.3321/j.issn:1001-7216.2000.02.001.
Gu X Y, Mei M T, Yan X L, Zheng S L, Lu Y G.Preliminary detection of quantitative trait loci for salt tolerance in rice[J].Chinese Journal of Rice Science,2000,14(2):65-70.
[15] 邢军,常汇琳,王敬国,刘化龙,孙健,郑洪亮,赵宏伟,邹德堂.盐、碱胁迫条件下水稻Na+、K+浓度的QTL分析[J].中国农业科学,2015,48(3):604-612. doi:10.3864/j.issn.0578-1752.2015.03.19.
Xing J, Chang H L, Wang J G, Liu H L, Sun J, Zheng H L, Zhao H W, Zou D T.QTL analysis of Na+ and K+ concentrations in Japonica rice under salt and alkaline stress[J].Scientia Agricultura Sinica,2015,48(3):604-612.
[16] Xing W,Zhao H W,Zou D T. Detection of main-effect and epistatic QTL for yield-related traits in rice under drought stress and normal conditions[J].Canadian Journal of Plant Science, 2014,94(4):633-641.doi:10.4141/cjps2013-297.
[17] Li H H,Ye G Y,Wang J K.A modified algorithm for the improvement of composite interval mapping[J].Genetics,2007,175(1):361-374.doi:10.1534/genetics.106.066811.
[18] McCouch S R, Cho Y G, Yang M, Paul E, Blinstrub M, Morishima H, Kinoshita T. Report on QTL nomenclature[J]. Rice Genet Newslet, 1997, 14: 11-13.
[19] 程立锐. 利用双向导入系和重组自交系研究水稻重要农艺性状的遗传规律[D].北京:中国农业科学院,2011.doi:10.7666/d.Y1932863.
Cheng L R.Study on the inheritance of important agronomic traits of rice by using two-way introduction system and recombinant inbred lines[D].Beijing:Chinese Academy of Agricultural Sciences,2011.
[20] 柴路. 利用高代回交导入系定位水稻全生育期耐盐QTL[D].北京:中国农业科学院,2013.
Chai L.Using high-generation backcross introduction to locate salt-tolerant QTLs in rice during the whole growth period[D].Beijing:Chinese Academy of Agricultural Sciences,2013.
[21] 曲英萍. 水稻耐盐碱性QTLs分析[D].北京:中国农业科学院,2007.
Qu Y P.Analysis of salt and alkaline QTLs in rice[D].Beijing:Chinese Academy of Agricultural Sciences,2013.
[22] 李宁. 水稻苗期耐碱QTL分析[D].哈尔滨:东北农业大学,2016.
Li N.QTL analysis of alkali tolerance in rice seedling[D].Harbin:Northeast Agricultural University,2016.
[23] 梁晶龙. 利用重组自交系群体的水稻耐盐/碱性QTL定位分析[D].重庆:重庆师范大学,2013.
Liang J L.Salt-tolerant/alkaline QTL mapping of rice using recombinant inbred lines[D].Chongqing:Chongqing Normal University,2013.
[24] 索艺宁,张春可,于乔乔,张恩源,谢冬微,冷月,王亮,孙健.盐、碱胁迫下水稻苗期根数和根长的QTL分析[J].华北农学报,2018,33(5):9-15.doi:10.7668/hbnxb.2018.05.002.
Suo Y N, Zhang C K, Yu Q Q, Zhang E Y, Xie D W, Leng Y, Wang L, Sun J.QTL analysis of root number and root length in rice seedling stage under salt and alkali stress[J].Acta Agriculturae Boreali-Sinica,2018,33(5):9-15.
[25] 程海涛,姜华,薛大伟,郭龙彪,曾大力,张光恒,钱前.水稻芽期与幼苗前期耐碱性状QTL定位[J].作物学报,2008,34(10):1719-1727.doi:10.3321/j.issn:0496-3490.2008.10.006.
Cheng H T, Jiang H, Xue D W, Guo L B, Zeng D L, Zhang G H, Qian Q.Mapping of QTLs underlying tolerance to alkali at germination and early seedling stages in rice[J].Acta Agronomica Sinica,2008,34(10):1719-1727.
[26] 陈剑,潘晓飚,谢留杰,黄善军.盐胁迫对水稻恢复系苗期生长生理的影响[J].浙江农业科学,2016,57(7):1039-1042.doi:10.16178/j.issn.0528-9017.20160725.
Chen J, Pan X B, Xie L J, Huang S J.Effects of salt stress on growth physiology of rice restorer lines at seedling stage[J].Journal of Zhejiang Agricultural Sciences,2016,57(7):1039-1042.
[27] 郑少玲, 严小龙. 盐胁迫下不同水稻基因型根中Na+和Cl-分布情况比较[J]. 华南农业大学学报, 1996, 17(4): 24-28.
Zheng S L, Yan X L. Distribution of Na+ and Cl- in the roots of different rice genotypes under salt stress[J].Journal of South China Agricultural University, 1996, 17(4): 24-28.
[28] Sharma S K. Mechanism of tolerance in rice varieties differing in sodicity tolerance[J].Plant and Soil,1986,93(1):141-145.doi:10.1007/BF02377155.
[29] Veldboom L R, Lee M, Woodman W L. Molecular-marker facilitated studies in an elite maize population: I. linkage analysis and determination of QTL for morphological traits[J]. Theoretical and Applied Genetics, 1994, 88: 7-16. doi:10.1007/BF00222387.
[30] Lin H X, Qian H R, Zhuang J Y, Lu J, Min S K, Xiong Z M, Huang N, Zheng K L. RFLP mapping of QTLs for yield and related characters in rice (Oryza sativa L.) [J]. Theoretical and Applied Genetics, 1996, 92: 920-927. doi:10.1007/BF00224031.
[31] Xiao J, Li J,Yuan L, Tanksley S D. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross[J]. Theoretical and Applied Genetics, 1996, 92: 230-244.doi:10.1007/BF00223380.
[32] 郑洪亮,刘博文,赵宏伟,王敬国,刘化龙,孙健,邢军,邹德堂.利用连锁和关联分析定位粳稻芽期及幼苗前期耐盐性QTL[J].中国水稻科学,2014,28(4):358-366.doi:10.3969/j.issn.1001-7216.2014.04.004.
Zheng H L, Liu B W, Zhao H W, Wang J G, Liu H L, Sun J, Xing J, Zou D T.Identification of QTLs for salt tolerance at the germination and early seedling stage using linkage and association analysis in japonica rice[J].Chinese Journal of Rice Science,2014,28(4):358-366.
[33] 邹德堂,张恩源,孙健,王敬国,刘化龙,郑洪亮.盐、碱条件下水稻剑叶形态相关性状QTL分析[J].东北农业大学学报,2017,48(11):9-17.doi:10.3969/j.issn.1005-9369.2017.11.002.
Zou D T, Zhang E Y, Sun J, Wang J G, Liu H L, Zheng H L.QTL analysis on characters related to flag leaf morphology of rice under salt and alkali conditions[J].Journal of Northeast Agricultural University,2017,48(11):9-17.
[34] Flowers T J,Koyama M L,Flowers S A,Sudhakar C,Singh K P,Yeo A R.QTL:their place in engineering tolerance of rice to salinity[J].Journal of Experimental Botany,2000,51(342):99-106.doi:10.1093/jexbot/51.342.99.
[35] Pandit A,Rai A,Bal S,Sinha S,Kumar V,Chauhan M,Gautam R K,Singh R,Sharma P C,Singh A K,Gaikwad K,Sharma T R,Mohapatra T,Singh N K.Combining QTL mapping and transcriptome profiling of bulked RILs for identification of functional polymorphism for salt tolerance genes in rices(Oryza sativa L.)[J].Molecular Genetics and Genomics,2010,284(2):121-136.doi:10.1007/s00438-010-0551-6.
[36] Mohammadi R,Mendioro M S,Diaz G Q,Gregorio G B,Singh R K.Mapping quantitative trait loci associated with yield and yield components under reproductive stage salinity stress in rice(Oryza sativa L.)[J].Journal of Genetics,2013:92:433-443.doi:10.1007/s12041-013-0285-4.
[37] Temnykh S, Declerck G, Lukashova A, Lipovich L, Cartinhour S, McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential[J].Genome Research,2001,11:1441-1452. doi:10.1101/gr.184001.
[38] Kurata N, Nagamura Y, Yamamoto K, Harushima Y, Sue N, Wu J, Antonio B A, Shomura A, Shimizu T, Lin S Y, Inoue T, Fukuda A, Shimano T, Kuboki Y, Toyama T, Miyamoto Y, Kirihara T, Hayasaka K, Miyao A, Monna L, Zhong H S, Tamura Y, Wang Z X, Momma T, Umehara Y, Yano M, Sasaki T, Minobe Y. A 300 kilobase interval genetic map of rice including 883 expressed sequences[J]. Nature Genetics, 1994, 8(4): 365-372.
[39] Ware D, Jaiswal P, Ni J J, Pan X, Chang K. Gramene: a resource for comparative grass genomics[J]. Nucleic Acid Research, 2001, 30(1): 103-105. doi:10.1093/nar/30.1.103.