胰岛素样生长因子家族(Insulin-like growth factor,IGF)包括3个配体、3个细胞膜受体、6个结合蛋白和许多其他相关蛋白[1]。在哺乳动物中,胰岛素调节细胞代谢,而胰岛素样生长因子1(IGF-1)是细胞生长的重要调控因子[2]。大部分循环的IGF-1是由肝脏产生的,生长激素可以调控肝脏表达IGF-1。然而,其他器官也可以产生自分泌和旁分泌的IGF-1,包括牛乳腺和肌肉[3]。胰岛素样生长因子1受体(IGF-1R)是一种受体-酪氨酸激酶,在对细胞生存和增殖至关重要的信号传导中起着至关重要的作用。研究显示,在多种肿瘤中IGF-1R出现上调表达,可能通过促进细胞增殖及抑制细胞凋亡等方式促进肿瘤的发生[4-5]。IGF-1R与原配体(IGF-1)结合引起一系列信号级联反应,从而激活磷酸肌醇3-激酶(PI3K)和mitogen活化蛋白激酶(MAPK)来促进细胞增殖[6],诱导凋亡蛋白的磷酸化和抑制,阻断细胞凋亡[7]。
IGF-1R基因多态性影响动物的生长,通过生物进化树分析大多数哺乳动物IGF-1R基因很保守[8]。为了进一步探讨IGF-1R基因的SNPs遗传多态性和连锁不平衡性,Lei等[9]针对鸡的18个SNPs研究,发现A17299834G SNP与鸡胴体的体质量显著相关,A17307750G、A17307494G SNP与早期生长性状相关。Wu等[10]利用PCR-RFLP方法检测边鸡IGF-1R基因2个多态位点,分别位于外显子2和3上,在8,14,16,18周时,具有AluI酶切位点AA基因型的母边鸡体质量高于AB基因型鸡(P<0.05);Hin1I酶切位点的CD基因型个体在6,8,10,12,14岁时体质量较高于CC基因型 (P<0.05或P<0.01)。宋姗姗等[11]采用直接测序法在大体型水貂银蓝水貂和小体型水貂美国短毛黑水貂中筛查IGF-1R基因21个外显子的SNPs,发现2个SNPs(c.207G>A、c.1782G>A),分别位于在外显子2和外显子11上,关联分析发现 c.1782G>A与美国短毛黑水貂体质量显著相关,且杂合型AA型为优势基因型。Szewczuk等[12]采用PCR-RFLP方法发现IGF-1R基因可作为研究水貂生长性状的一个候选基因,该基因的多态性及基因型效应在不同种群水貂中不同,因此,本研究下一步可增加种群数量并对IGF-1R基因的多态性进行分析,并与水貂生长性状进行关联分析,为培育体型大的水貂和水貂的育种工作奠定理论基础。
1 材料和方法
1.1 试验材料
以2017年12月打皮期3种群水貂共235只为研究对象,其中包括国外引进品种红眼白水貂118只,咖啡水貂50只,国内培育品种金州黑水貂67只,取自大连名威貂业,打皮时心脏采血于5 mL真空采血管中,统计水貂个体测量体尺和体质量数据。
1.2 血液全基因组DNA提取
血液DNA的提取采用传统的酚氯仿法[13]。应用琼脂糖凝胶电泳和紫外分光光度计方法检测DNA的完整性、浓度及纯度,将其质量浓度稀释成20 ng/μL,-20 ℃保存[14]。
1.3 PCR扩增及测序分析
水貂IGF-1R序列外显子2和外显子11特异性引物设计参照参考文献[11],由吉林省库美生物科技有限公司合成。以提取全基因DNA为模板,进行PCR扩增,PCR反应体系50 μL:模板DNA 1.0 μL、上下游引物(10 μmol/L)各1 μL,TransStart KD Plus DNA Polymerase 1 μL、5×TransStart KD Plus Buffer 10 μL,2.5 mmol/L dNTPs 4 μL,灭菌超纯水32 μL。反应条件为:95 ℃预变性5 min; 94 ℃变性30 s,55 ℃退火30 s,72 ℃延伸20 s,循环数为35,72 ℃后延伸10 min。采用1.2%琼脂糖凝胶电泳检测PCR产物,目的片段经切胶纯化后,16 ℃过夜连接到pEASY-Blunt Simple Cloning Kit载体上,转化到DH5α感受态细胞中,过夜培养后挑平板上单菌落,经菌液PCR鉴定为阳性样品后,送北京六合华大基因科技有限公司测序。
1.4 IGF-1R SNPs位点查询及其基因型的判定
采用BioEdit软件Clustal W Multiple alignment程序进行IGF-1R基因序列的比对和分析,筛查SNPs位点;采用SeqMan软件查看序列对应的峰图判定SNPs的基因型,单一峰为纯合基因型,套峰为杂合基因型。
1.5 基因型、等位基因频率及SNPs位点与生长性状的关联分析
采用Popgene1.32软件分析基因型频率和等位基因频率,并利用SAS 9.1中的PROC GLM程序进行SNPs位点与生长性状的关联分析(SAS Institute, Inc, Cary, NC, USA)。
2 结果与分析
2.1 PCR扩增IGF-1R基因结果
利用设计的特异性引物对不同个体水貂IGF-1R基因进行扩增,扩增产物见图1。2对特异性引物扩增片段为180,236 bp,与预期设计扩增长度相一致,进行克隆反应。
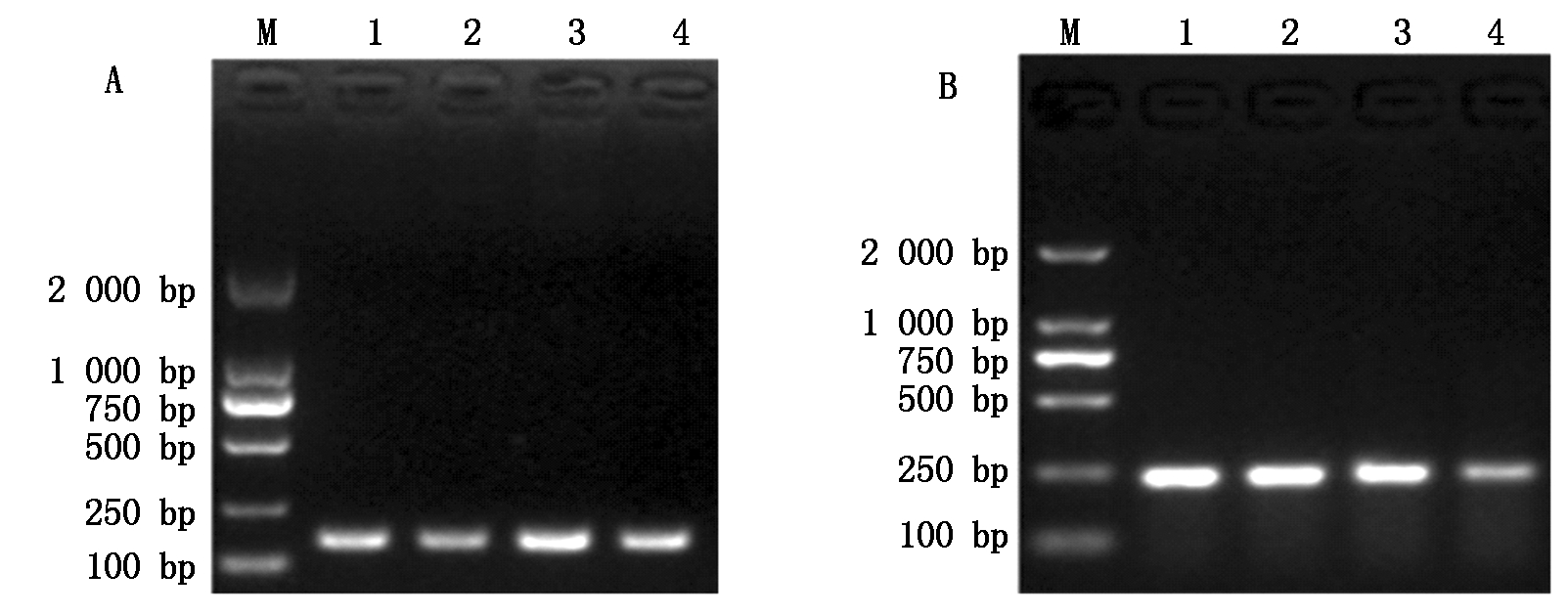
A.1-4. 引物2 PCR产物;B.1-4.引物11 PCR产物;M.DNA Marker DL2000.
A.1-4.The PCR products of primer 2;B.1-4.The PCR products of primer 11;M.DNA Marker DL2000.
图1 PCR扩增水貂IGF-1R基因电泳图谱
Fig.1 Electrophoresis of the products of IGF-1R gene in mink
2.2 水貂IGF-1R基因鉴定
对纯化后的2段PCR产物进行克隆(图2)、测序,获得片段长度分别为180,236 bp,通过Blast比对发现,序列一和序列二与NCBI公布的雪貂同源性达到99%,97%,可认定克隆得到的2条序列为水貂IGF-1R基因序列。
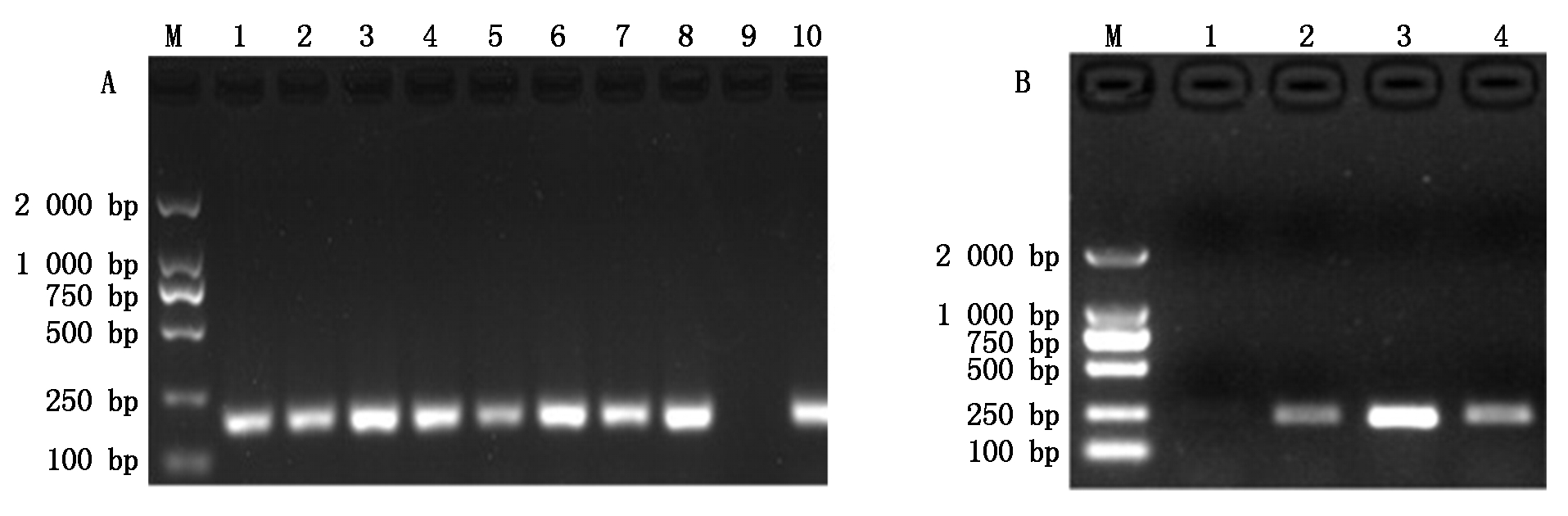
A.1-10.引物2扩增的PCR片段;B.1-4.引物11扩增的PCR片段;M.DNA Marker DL2000。
A.1-10. The PCR result of bacteria colony of primer 2; B. 1-4. The PCR result of bacteria colony of primer 11; M. DNA Marker DL2000.
图2 菌液PCR鉴定电泳图谱
Fig.2 PCR result of bacteria colony
2.3 SNPs位点筛查及基因型判定
参照雪貂IGF-1R基因序列信息分析2对引物的测序结果,获得长度为180,230 bp的外显子序列。采用BioEdit 7.0软件对3种群水貂的测序结果进行拼接比对,结果显示,红眼白水貂和金州黑水貂在引物2扩增的序列检测到c.207G>A,在引物11扩增的序列检测到c.1782G>A;咖啡水貂引物2扩增的序列检测到c.207G>A和c.218T>A,在引物11扩增的序列检测到c.1782G>A。其中红眼白水貂、金州黑水貂和咖啡水貂c.207G>A和c.1782G>A位点都属于无义突变,氨基酸未发生改变。咖啡水貂c.218T>A位点属于有义突变,氨基酸发生改变,脯氨酸变为精氨酸。红眼白水貂c.207G>A检测到2个基因型;c.1782G>A检测到3个基因型;金州黑水貂c.207G>A检测到3个基因型;c.1782G>A检测到3个基因型;咖啡水貂c.207G>A检测到2个基因型,c.218T>A检测到2个基因型,c.1782G>A检测到3个基因型(图3-9)。
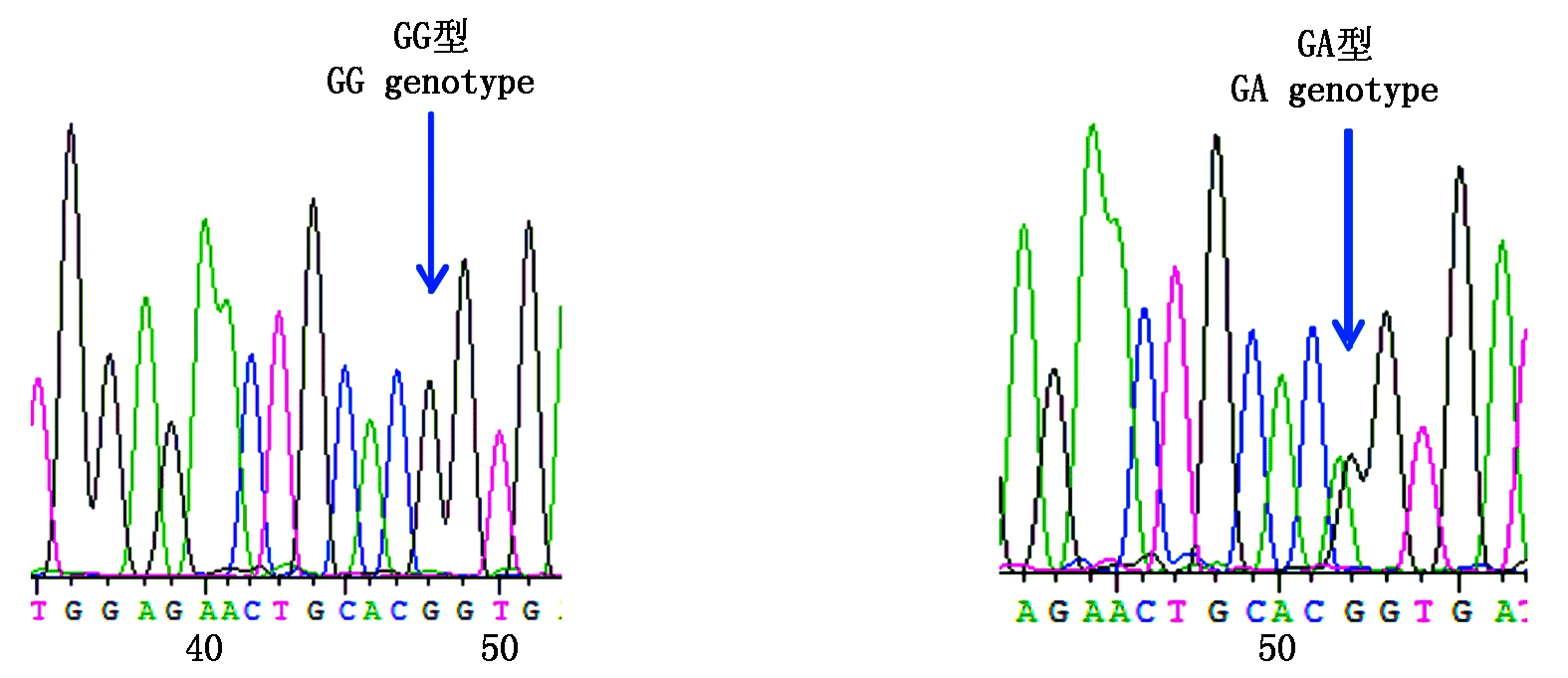
图3 红眼白水貂c.207G>A变异位点峰图及分型图
Fig.3 Nucleotide sequence variation peak figure and genotyped chart of c.207G>A in red-eye white mink

图4 红眼白水貂c.1782G>A变异位点峰图及分型图
Fig.4 Nucleotide sequence variation peak figure and genotyped chart of c.1782G>A in red-eye white mink
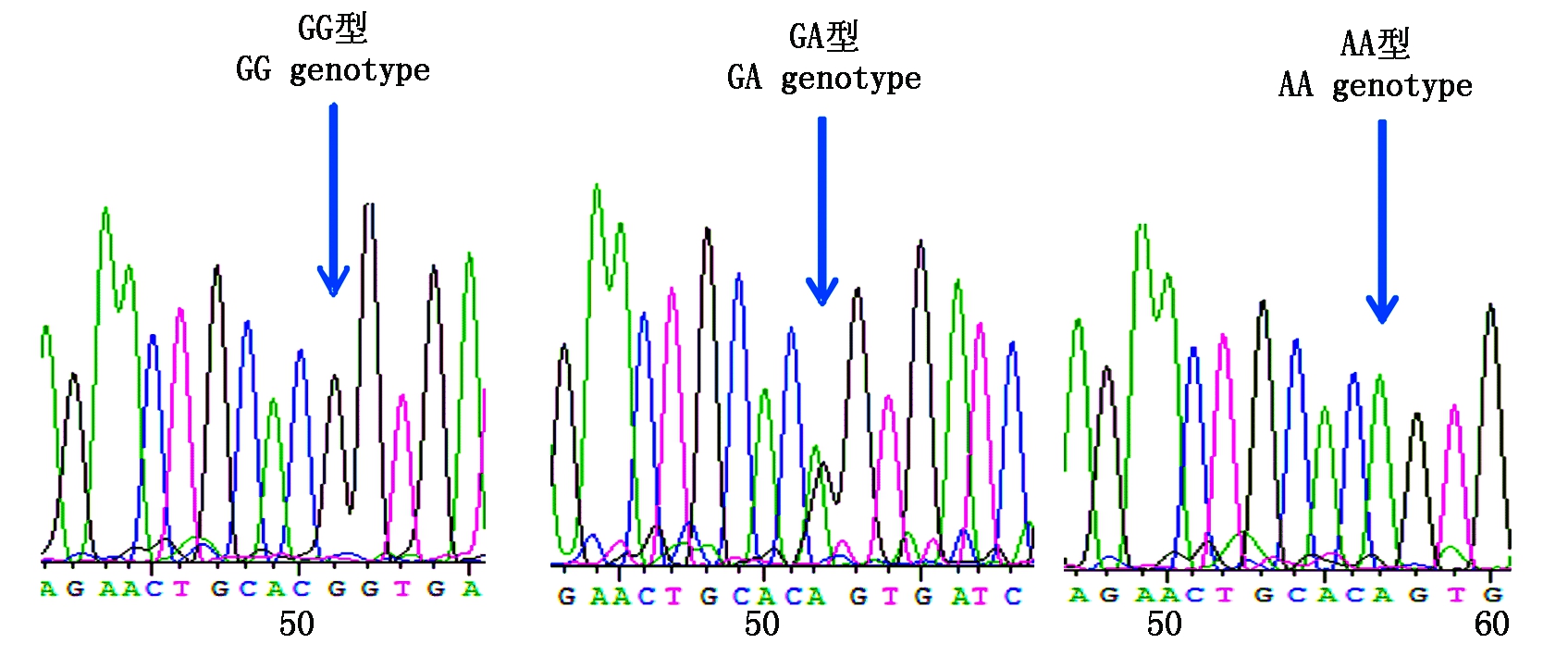
图5 金州黑水貂c.207G>A变异位点峰图及分型图
Fig.5 Nucleotide sequence variation peak figure and genotyped chart of c.207G>A in Jinzhou black mink
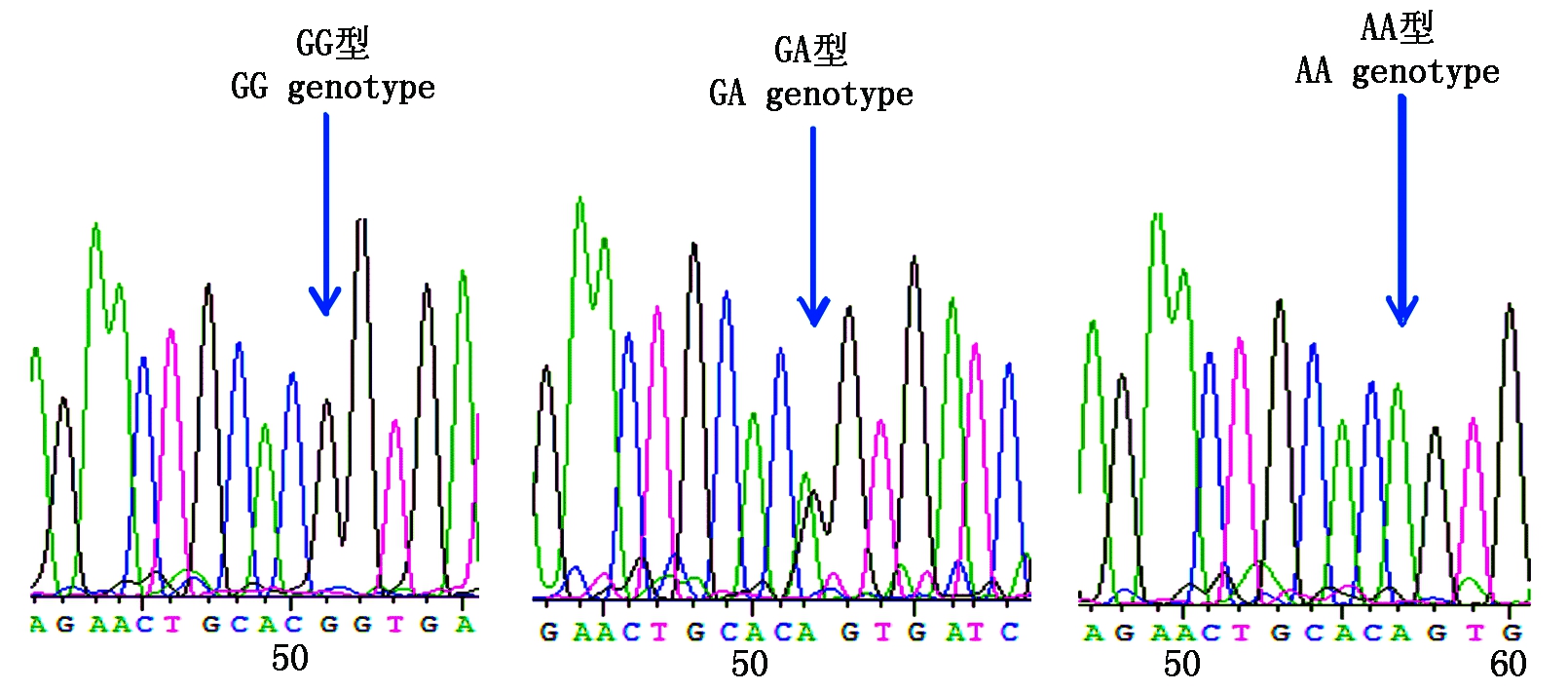
图6 金州黑水貂c.1782G>A变异位点峰图及分型图
Fig.6 Nucleotide sequence variation peak figure and genotyped chart of c.1782G>A in Jinzhou black mink
2.4 SNPs位点的群体遗传结构分析
利用PopGene32分析各SNPs位点的遗传参数见表1,2。红眼白水貂、咖啡水貂c.207G>A位点属于低度多态(P<0.25),其他位点在3种水貂中属于中度多态(0.25<P<0.5)。卡方检验显示,3个SNPs位点的基因型分布符合Hardy-Weinberg平衡状态。
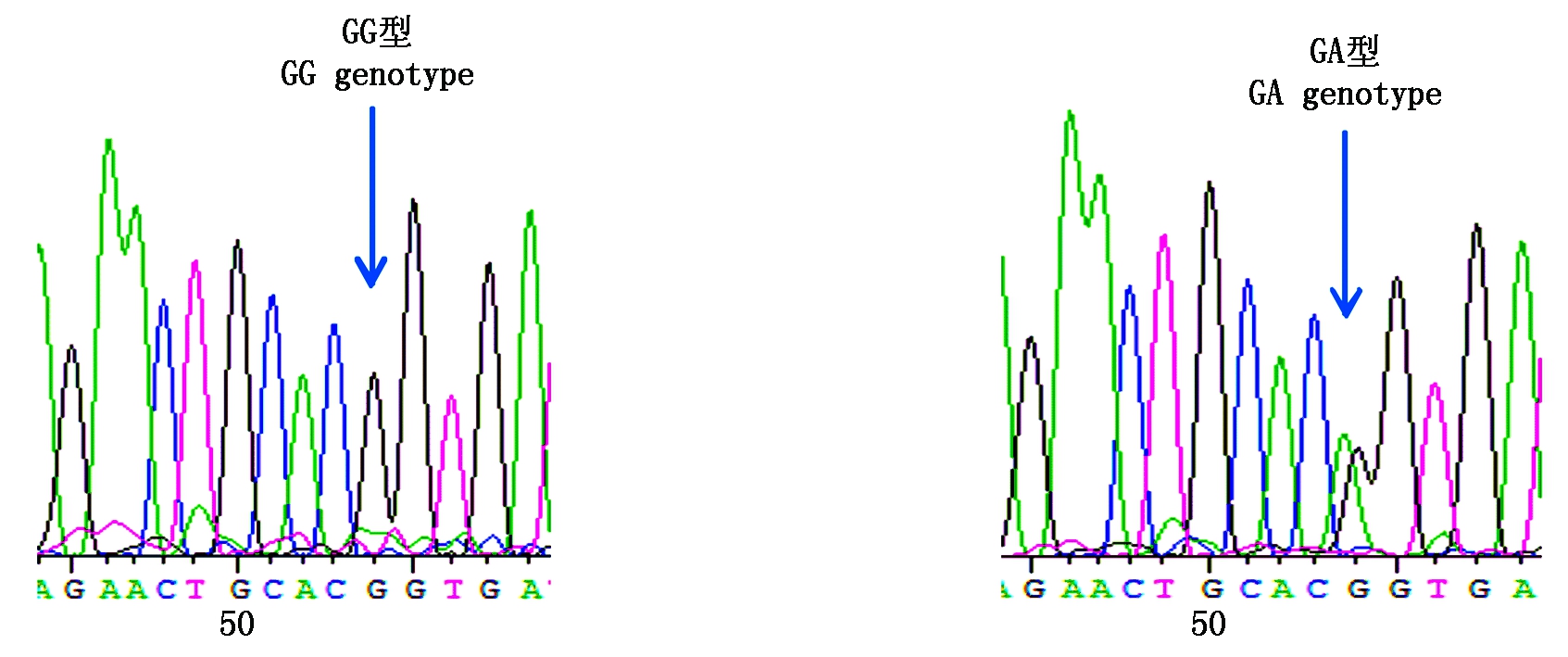
图7 咖啡水貂c.207G>A变异位点峰图及分型图
Fig.7 Nucleotide sequence variation peak figure and genotyped chart of c.207G>A in coffee mink
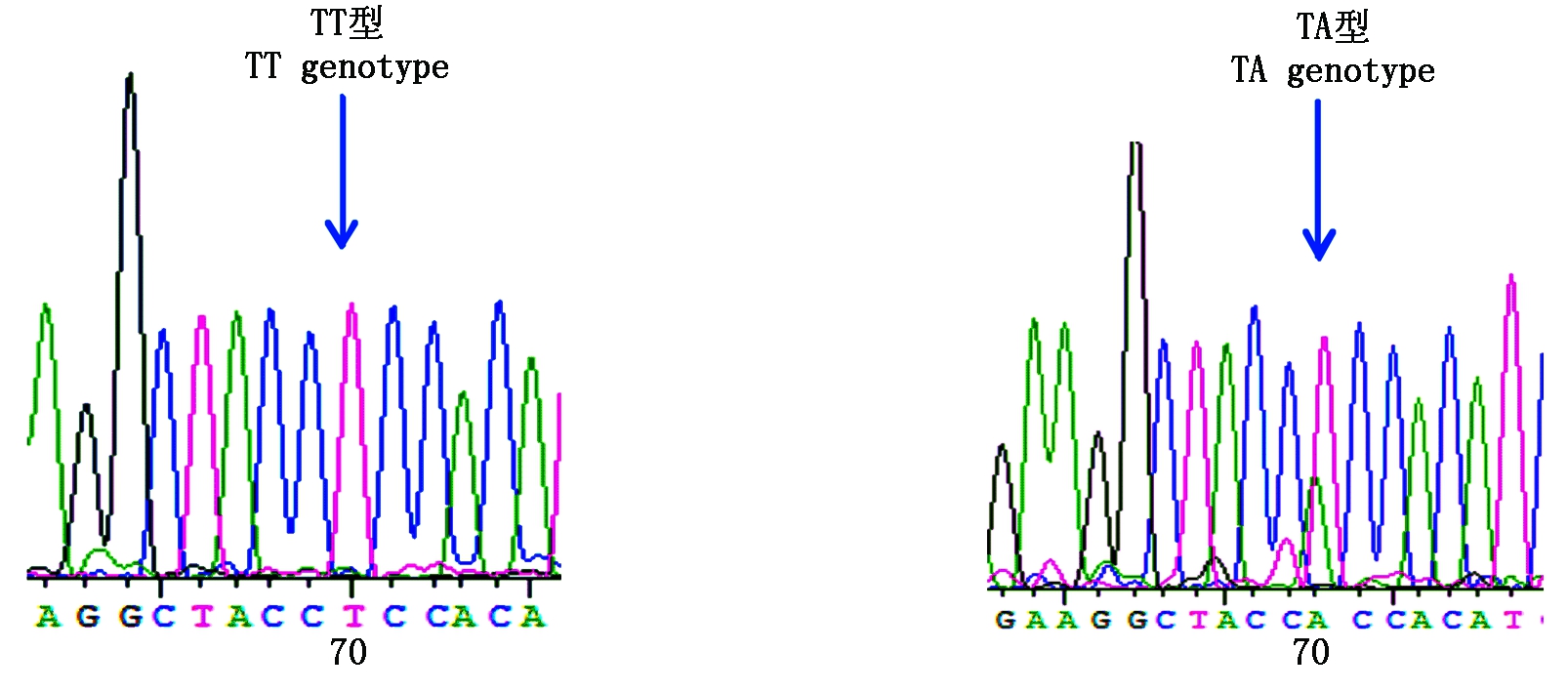
图8 咖啡水貂c.218T>A变异位点峰图及分型图
Fig.8 Nucleotide sequence variation peak figure and genotyped chart of c.218T>A in coffee mink
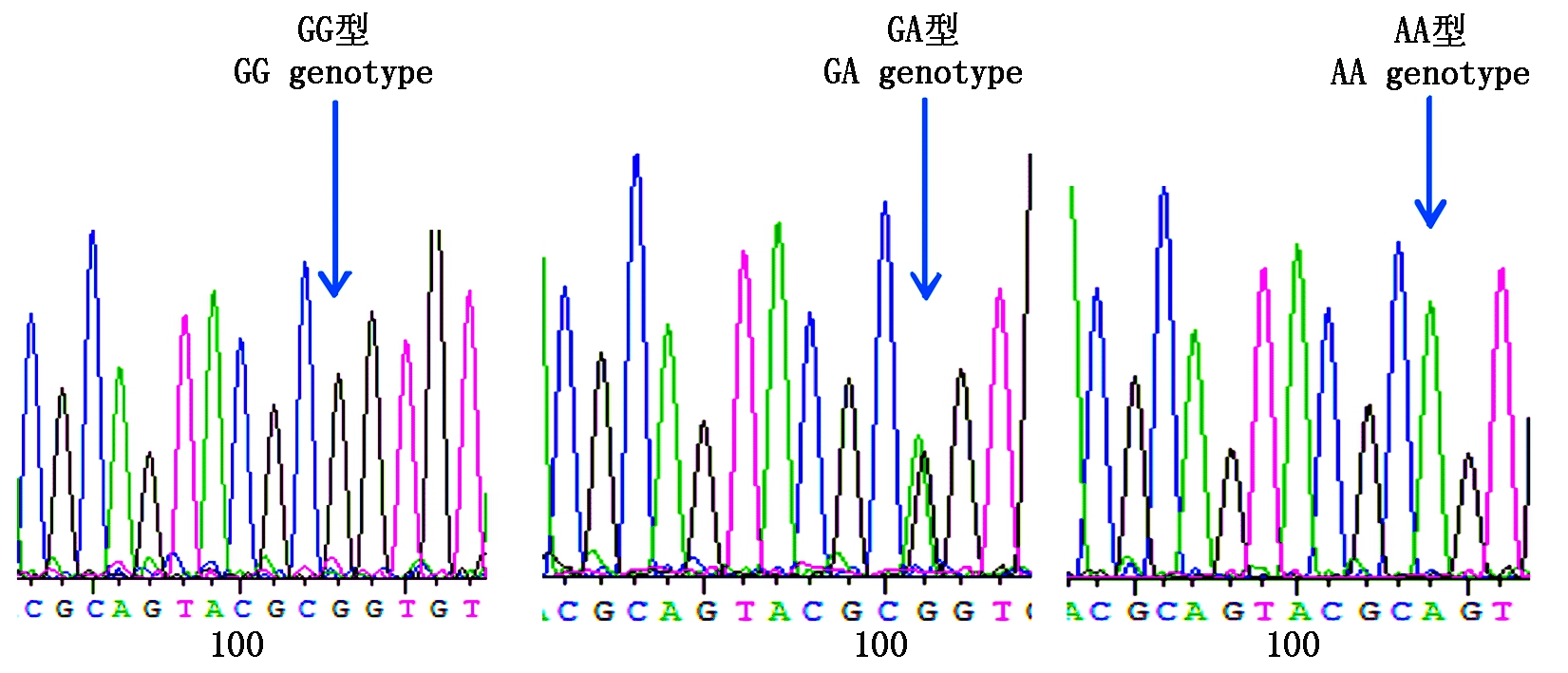
图9 咖啡水貂c.1782G>A变异位点峰图及分型图
Fig.9 Nucleotide sequence variation peak figure and genotyped chart of c.1782G>A in coffee mink
表1 SNPs位点的基因型和等位基因频率
Tab.1 Genotypic and allelic frequencies of SNPs
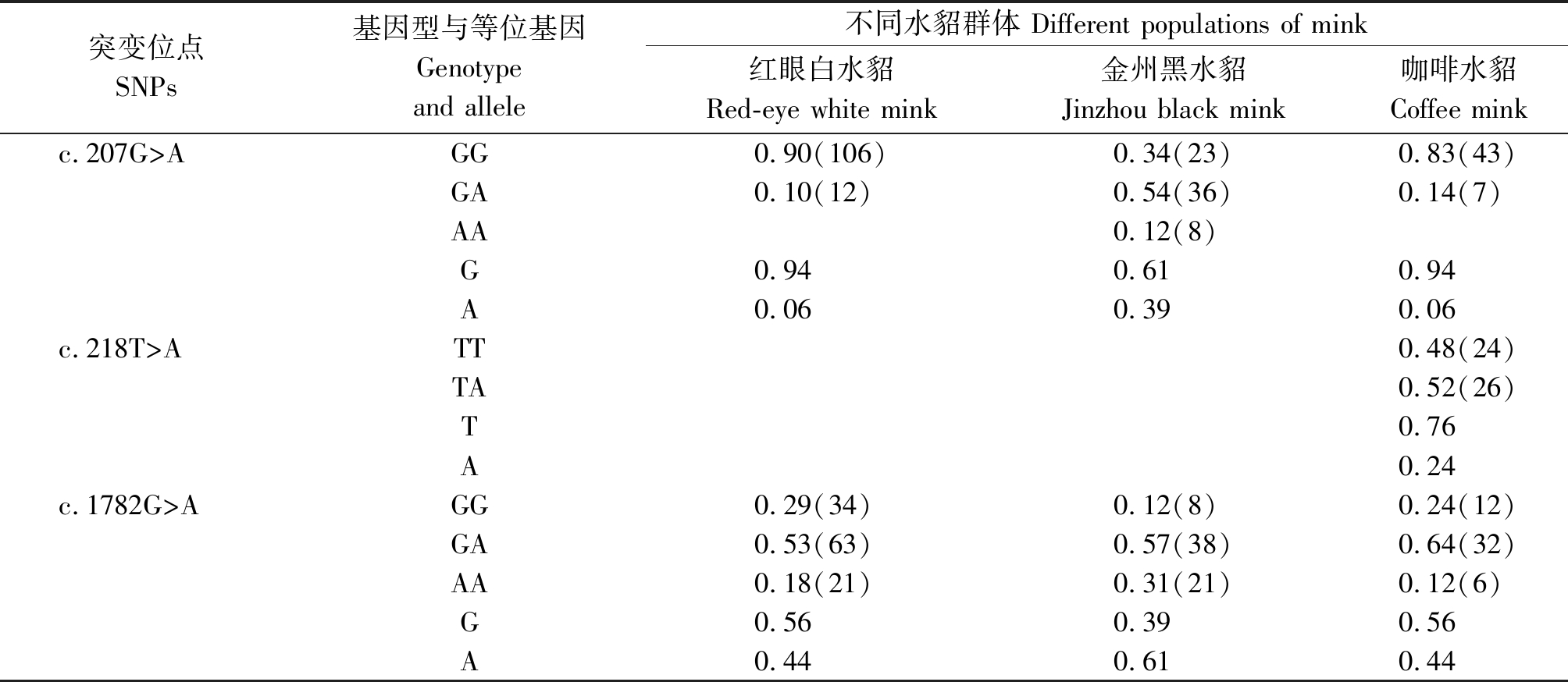
突变位点SNPs基因型与等位基因Genotype and allele不同水貂群体 Different populations of mink红眼白水貂Red-eye white mink金州黑水貂Jinzhou black mink咖啡水貂Coffee minkc.207G>AGG0.90(106)0.34(23)0.83(43)GA0.10(12)0.54(36)0.14(7)AA0.12(8)G0.940.610.94A0.060.390.06c.218T>ATT0.48(24)TA0.52(26)T0.76A0.24c.1782G>AGG0.29(34)0.12(8)0.24(12)GA0.53(63)0.57(38)0.64(32)AA0.18(21)0.31(21)0.12(6)G0.560.390.56A0.440.610.44
表2 等位基因多态性群体遗传结构分析
Tab.2 Population genetic structure based on allelic polymorphism data
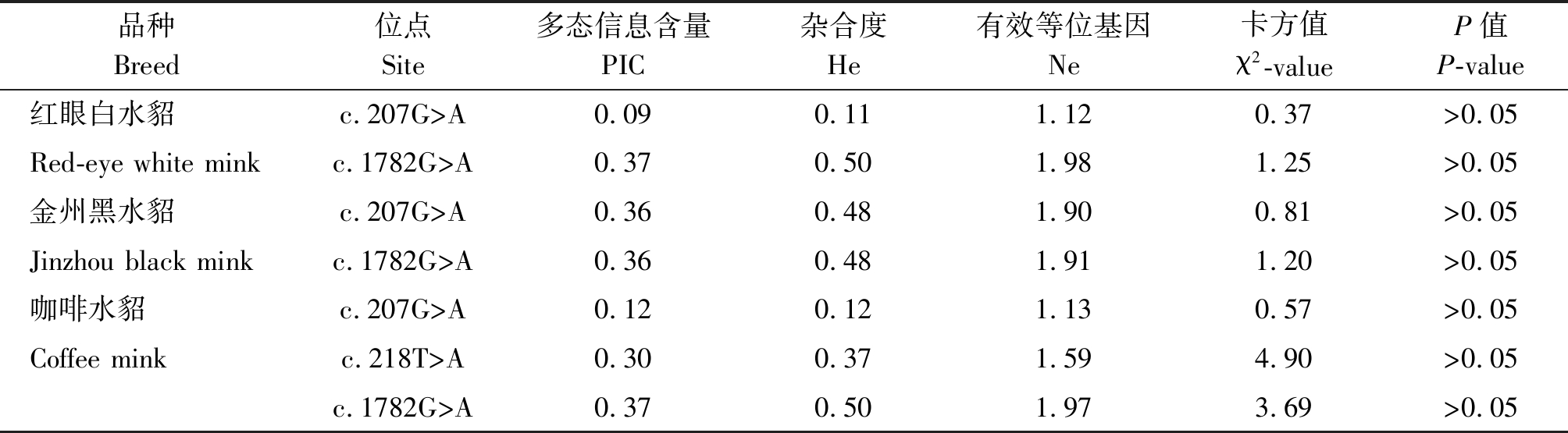
品种Breed位点Site多态信息含量PIC杂合度He有效等位基因Ne卡方值χ2-valueP值P-value红眼白水貂c.207G>A0.090.111.120.37>0.05Red-eye white minkc.1782G>A0.370.501.981.25>0.05金州黑水貂c.207G>A0.360.481.900.81>0.05Jinzhou black minkc.1782G>A0.360.481.911.20>0.05咖啡水貂c.207G>A0.120.121.130.57>0.05Coffee minkc.218T>A0.300.371.594.90>0.05c.1782G>A0.370.501.973.69>0.05
2.5 IGF-1R基因SNPs位点与水貂生长性状的关联分析
SNPs位点基因型与生长性状的关联分析结果见表3。c.207G>A与金州黑水貂和咖啡水貂体长性状显著相关(P<0.05),与红眼白水貂体长性状不显著相关,其中金州黑水貂野生型GG基因型个体体长比突变杂合GA基因型和突变纯合AA型的个体体长长;咖啡水貂野生型GG基因型个体体长长于突变杂合GA基因型。c.1782G>A与咖啡水貂的体长显著相关(P<0.05),与咖啡水貂的体质量呈现差异极显著相关(P<0.01),其中优势基因型为突变杂合GA基因型,其体长、体质量都显著高于其他2个基因型个体。
合并基因型与水貂生长性状关联分析见表4。金州黑水貂合并基因型GGGA个体和GGAA个体的体长与GAGA个体体长差异极显著(P<0.01),基因型GGGA个体和GGAA个体的体长长于GAGA个体体长;金州黑水貂合并基因型GGGA个体体质量与GAGA个体差异显著(P<0.05),GGGA个体体质量高于GAGA基因型个体。试验可推断得出合并基因型GGGA和GGAA可能是影响金州黑水貂体长体质量的有利基因型。
表3 c.207G>A、c.218T>A 和c.1782G>A位点不同基因型与3种群水貂生长性状关联分析
Tab.3 Correlation between different genotypes and growth traits of c.207G>A, c.218T>A and c.1782G>A sites in three groups
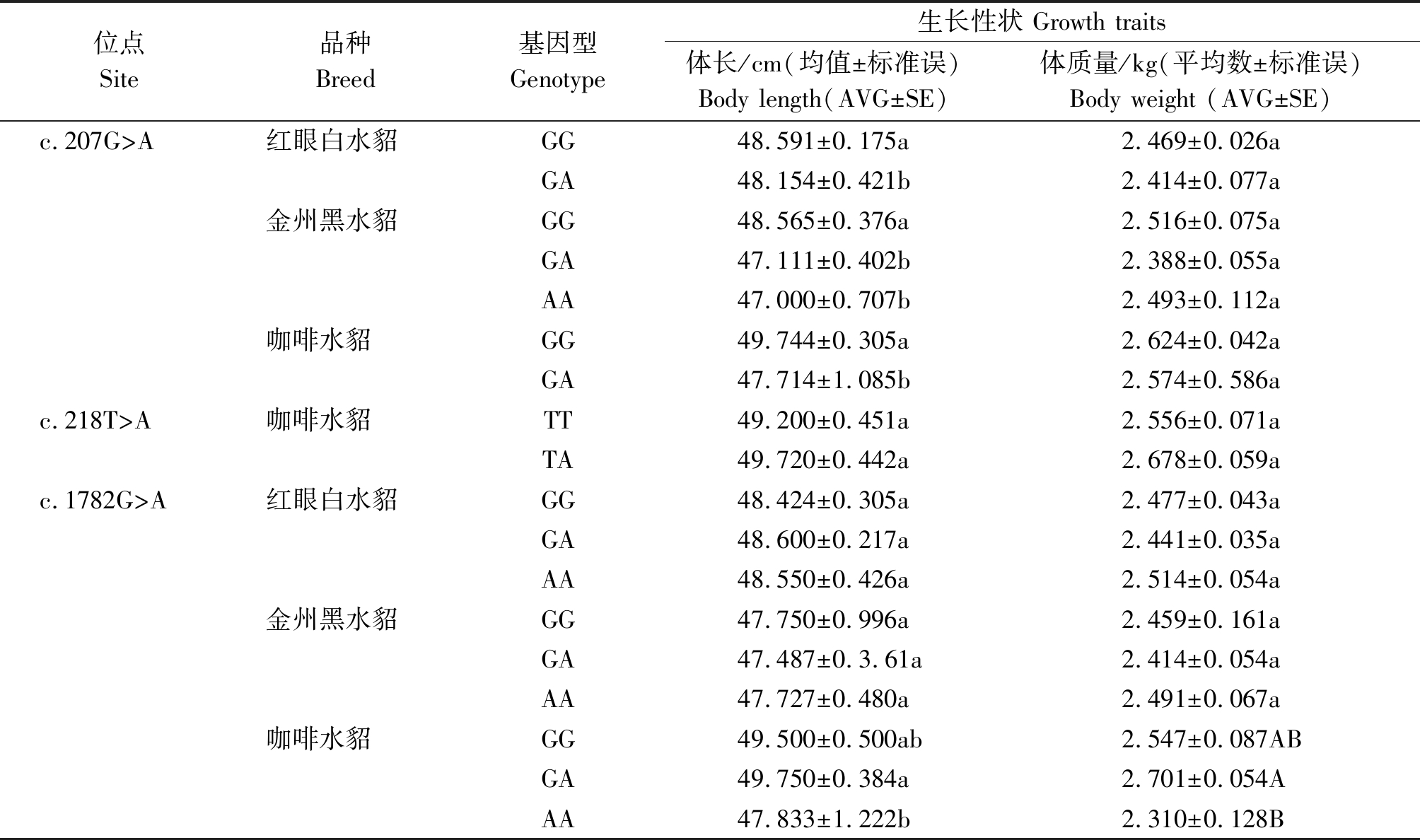
位点Site品种Breed基因型 Genotype生长性状 Growth traits体长/cm(均值±标准误)Body length(AVG±SE)体质量/kg(平均数±标准误)Body weight (AVG±SE)c.207G>A红眼白水貂GG48.591±0.175a2.469±0.026aGA48.154±0.421b2.414±0.077a金州黑水貂GG48.565±0.376a2.516±0.075aGA47.111±0.402b2.388±0.055aAA47.000±0.707b2.493±0.112a咖啡水貂GG49.744±0.305a2.624±0.042aGA47.714±1.085b2.574±0.586ac.218T>A咖啡水貂TT49.200±0.451a2.556±0.071aTA49.720±0.442a2.678±0.059ac.1782G>A红眼白水貂GG48.424±0.305a2.477±0.043aGA48.600±0.217a2.441±0.035aAA48.550±0.426a2.514±0.054a金州黑水貂GG47.750±0.996a2.459±0.161aGA47.487±0.3.61a2.414±0.054aAA47.727±0.480a2.491±0.067a咖啡水貂GG 49.500±0.500ab 2.547±0.087ABGA49.750±0.384a 2.701±0.054AAA47.833±1.222b 2.310±0.128B
注:同列不同小写字母表示差异显著(P<0.05);同列不同大写字母表示差异极显著(P<0.01)。表4同。
Note: Different small letters in each column mean significant difference(P<0.05); while capital letters mean extremely significant difference(P<0.01).The same as Tab.4.
表4 c.207G>A、c.218T>A 和c.1782G>A位点合并基因型与3种群水貂生长性状关联分析
Tab.4 Correlation between different combination genotypes and growth traits of c.207G>A, c.218T>A and c.1782G>A sites in three groups
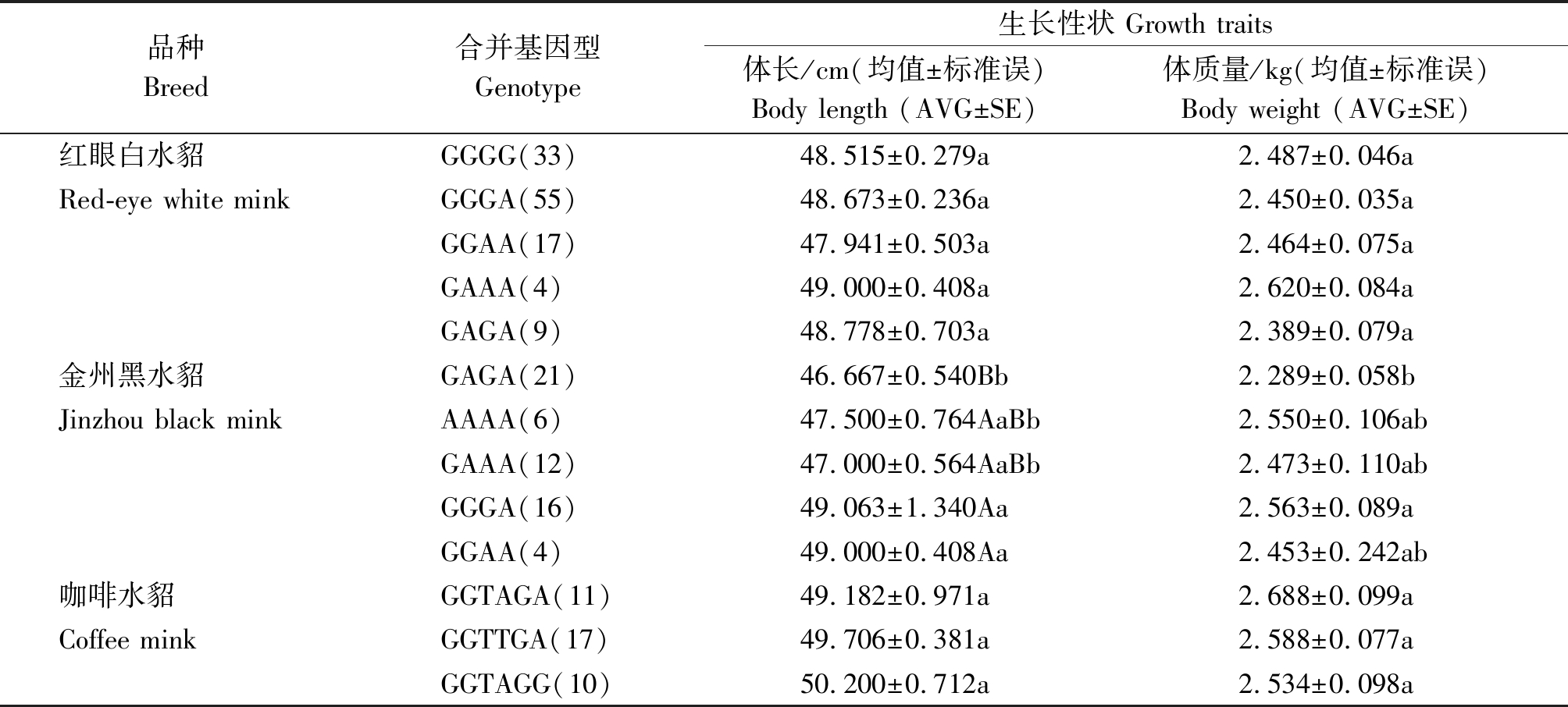
品种Breed合并基因型Genotype生长性状 Growth traits体长/cm(均值±标准误)Body length (AVG±SE)体质量/kg(均值±标准误)Body weight (AVG±SE)红眼白水貂GGGG(33)48.515±0.279a2.487±0.046aRed-eye white minkGGGA(55)48.673±0.236a2.450±0.035aGGAA(17)47.941±0.503a2.464±0.075aGAAA(4)49.000±0.408a2.620±0.084aGAGA(9)48.778±0.703a2.389±0.079a金州黑水貂GAGA(21) 46.667±0.540Bb2.289±0.058bJinzhou black minkAAAA(6) 47.500±0.764AaBb 2.550±0.106abGAAA(12) 47.000±0.564AaBb 2.473±0.110abGGGA(16) 49.063±1.340Aa2.563±0.089aGGAA(4) 49.000±0.408Aa 2.453±0.242ab咖啡水貂GGTAGA(11)49.182±0.971a2.688±0.099aCoffee minkGGTTGA(17)49.706±0.381a2.588±0.077aGGTAGG(10)50.200±0.712a2.534±0.098a
3 结论与讨论
经典育种方案通过表型筛选和利用系谱信息实现遗传改良。这些项目提高了大多数现有作物品种和动物品种的生产力。然而,分子辅助育种的进展可以提高育种者选择最佳基因组合的优良表型的效率。与数量性状相关的基因组区域(QTL)可以使用多态DNA标记绘制,如微卫星或SNPs(单核苷酸多态性)[15]。这种关联可以加速育种方案的结果,使优等个体的早期识别具有更高的精度[16]。
动物的生长轴为GH-GHR-IGF-1-IGF-1R,IGF-1R基因作为生长轴的终端与IGF-1结合发挥生物学作用,它们形成的复合体能促进IGF-1R酪氨酸残基自身磷酸化,信号级联激活磷酸肌醇3-激酶(PI3K)和Ras蛋白信号通路从而阻断细胞凋亡[17],调控GH的分泌、影响机体生长发育,促进细胞凋亡、增殖和分化[18]。因此,进一步对水貂IGF-1R基因的生物学功能进行研究显得尤为重要。
先前在大体型银蓝水貂和小体型美国短毛黑两种群水貂研究IGF-1R基因,在外显子2和外显子11中发现2个SNPs,性状关联分析发现c.1782G>A与美国短毛黑水貂体质量显著相关,且AA型为优势基因型[19]。本研究中,在红眼白水貂、金州黑水貂和咖啡水貂3种群水貂中发现3个SNPs,其中2个SNPs与先前研究相同,氨基酸分析发现两突变位点均为同义突变,其氨基酸并未发生改变,但有研究表明,同一氨基酸的密码子有数量不等的tRNA,蛋白质表达的速度和数量将不同[20]。关联分析发现c.207G>A与金州黑水貂和咖啡水貂体长性状显著相关(P<0.05),与红眼白水貂体长性状不显著相关,c.1782G>A与咖啡水貂的体长显著相关,GA基因型个体体长与AA基因型个体体长差异显著,且GA基因型个体体质量与AA基因型个体体质量差异极显著。同时在咖啡水貂中发现一个新的SNPs:c.218T>A,该位点进行氨基酸分析发现位点属于有义突变,氨基酸发生改变,脯氨酸变为精氨酸。但进一步的性状关联分析发现该位点与水貂体长体质量性状并无显著相关性,本研究由于样品量较少,该结论需要进一步扩大样本量。试验表明,c.207G>A位点GG基因型是水貂体长的优势基因型,进一步进行了合并基因型与水貂生长性状关联分析,GGGA基因型和GGAA可能是影响金州黑水貂体长体质量的有利基因型。
综上,IGF-1R基因可作为研究水貂生长性状的一个候选基因,该基因的多态性及基因型效应在不同种群水貂中不同。应增加对IGF-1R基因的多态性及功能的研究,并分析其在水貂发育机制早中期的影响,及其该基因的生物学功能。揭示该候选基因在水貂生长发育过程中的作用机制。
[1] Pozios K C, Ding J, Degger B, Upton Z, Duan C. IGFs stimulate zebrafish cell proliferation by activating MAP kinase and PI3-kinase-signaling pathways [J]. American Journal of Physiology Regulatory, Integrative and Comparative Physiology, 2001, 280(4): 1230-1239. doi: 10.1152/ajpregu. 2001. 280. 4. R1230.
[2] Ullrich A, Gray A, Tam A W, Yang-Feng T, Tsubokawa M, Collins C W, Henzel W, Bon Le T, Kathuria S, Chen E S.Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity [J]. The EMBO Journal, 1986, 5(10): 2503-2512. doi: 10.1002/j.1460-2075.1986.tb04528.x.
[3] Plath-Gabler A, Gabler C, Sinowatz F, Berisha B, Schams D. The expression of the IGF family and GH receptor in the bovine mammary gland [J]. Journal of Endocrinology, 2001, 168(1): 39-48. doi: 10.1677/joe.0.1680039.
[4] Lovly C M, McDonald N T, Chen H O, Ortiz-Cuaran S, Heukamp L C, Yan Y J, Florin A, Ozretic' L, Lim D, Wang L, Chen Z, Chen X, Lu P C, Paik P K, Shen R L, Jin H L, Buettner R, Ansén S, Perner S, Brockmann M, Bos M, Wolf J, Gardizi M, Wright G M, Solomon B, Russell P A, Rogers T M, Suehara Y, Red-Brewer M, Tieu R, de Stanchina E, Wang Q G, Zhao Z M, Johnson D H, Horn L, Wong K K, Thomas R K, Ladanyi M, Pao W. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer [J]. Nature Medicine, 2014, 20(9):1027-1034. doi:10.1038/nm.3667.
[5] Aleksic T, Verrill C, Bryant R J, Han C, Worrall A R, Brureau L, Larré S, Higgins G S, Fazal F, Sabbagh A, Haider S, Buffa F M, Cole D, Macaulay V M.IGF-1R associates with adverse outcomes after radical radiotherapy for prostate cancer [J]. British Journal of Cancer, 2017, 117(11): 1600-1606. doi: 10. 1038/bjc. 2017. 337.
[6] 唐勇民. VEGF、IGF-1及其受体在子宫内膜癌中的表达及意义[J]. 现代实用医学, 2015, 27(7): 900-901, 907. doi: 10.3969/j.issn.1671-0800.2015.07.034.
Tang Y M. The expression of VEGF, IGF-1 and their receptors in endometrial carcinoma [J]. Modern Practical Medicine, 2015, 27(7): 900-901, 907.
[7] Galvan V, Logvinova A, Sperandio S, Ichijo H, Bredesen D E.Type 1 insulin-like growth factor receptor(IGF-IR)signaling inhibits apoptosis signal-regulating kinase 1(ASK1)[J]. The Journal of Biological Chemistry, 2003, 278(15): 13325-13332. doi: 10. 1074/jbc. M211398200.
[8] Perrot V, Moiseeva E B, Gozes Y, Chan S J, Ingleton P, Funkenstein B. Ontogeny of the insulin-like growth factor system(IGF-I, IGF-II, and IGF-1R)in gilthead seabream(Sparusaurata): Expression and cellular localization [J]. General and Comparative Endocrinology, 1999, 116(3): 445-460. doi:10.1006/gcen.1999.7337.
[9] Lei M M, Peng X, Zhou M, Luo C L, Nie Q H, Zhang X Q. Polymorphisms of the IGFIR gene and their genetic effects on chicken early growth and carcass traits [J]. BMC Genetics, 2008, 9(1): 70. doi: 10. 1006/gcen. 1999.7337.
[10] Wu P F, Wang D, Jin C F, Zhang X Q, Wu H Q, Zhang L, Ding F X, Xie K Z, Zhang G X. Polymorphisms of AluⅠ and HinⅡ loci of the IGF-1R gene and their genetic effects on growth traits in Bian chickens [J]. Genetics and Molecular Research, 2017, 16(2). doi: 10.4238/gmr.16029619.
[11] 宋姗姗, 王雷, 宋兴超, 邢秀梅, 杨福合. 水貂IGF-1R基因SNPs筛查及其与生长性状的关联分析 [J]. 吉林农业大学学报, 2016, 38(3): 350-356. doi:10.13327/j.jjlau.2015.2779.
Song S S, Wang L, Song X C, Xing X M, Yang F G. SNPs detection of IGF-1R gene and its correlation with growth trait in mink [J]. Journal of Jilin Agricultural University, 2016, 38(3): 350-356.
[12] Szewczuk M, Zych S, Wójcik J, Czerniawska-Piatkowska E. Association of two SNPs in the coding region of the insulin-like growth factor 1 receptor(IGF1R)gene with growth-related traits in Angus cattle[J].Journal of Applied Genetics, 2013, 54(3): 305-308. doi: 10. 1007/s13353-013-0155-z.
[13] Braakman R B, Bezstarosti K, Sieuwerts A M, De Weerd V, Van Galen A M, Stingl C, Luider T M, Timmermans M, Smid M, Martens J, Foekens J A, Demmers J, Umar A. Integrative analysis of genomics and proteomics data on clinical breast cancer tissue specimens extracted with acid guanidinium thiocyanate-phenol-chloroform [J]. Journal of Proteome Reseach, 2015, 14(3): 1627-1636. doi:10.1021/acs.jproteome.5b00046.
[14] 宋桃伟, 蔡惠芬, 罗卫星, 刘若余, 张依裕, 孙岩岩, 刘彬. 两个贵州山羊品种GHSR和GHRL基因遗传变异及其与生长性状的关联性[J]. 中国农业科学, 2015, 48(1):140-153. doi:10.3864/j.issn.0578-1752.2015.01.14.
Song T W, Cai H F, Luo W X, Liu R Y, Zhang Y Y, Sun Y Y, Liu B. Association of GHSR and GHRL gene genetic variation with growth traits in two Guizhou goat breeds [J]. Chinese Academy of Agricultural Sciences, 2015, 48(1): 140-153.
[15] Yoon S S, Kim E K, Lee W J. Functional genomic and metagenomic approaches to understanding gut microbiota-animal mutualism [J]. Current Opinion in Microbiology, 2015,24:38-46.doi:10.1016/j.mib.2015.01.007.
[16] Liu Z J, Cordes J F.DNA marker technologies and their applications in aquaculture genetics [J]. Aquaculture, 2004, 238(1/4): 1-37.doi:10.1016/j.aquaculture.2004.05.027.
[17] Ren G X, Ali T, Chen W, Han D D, Zhang L M, Gu X L, Zhang S Y, Ding L D, Fanning S, Han B. The role of selenium in insulin-like growth factor I receptor(IGF-IR)expression and regulation of apoptosis in mouse osteoblasts [J]. Chemosphere, 2016, 144: 2158-2164. doi: 10. 1016/j. chemosphere. 2015. 11. 003.
[18] Freier S, Weiss O, Eran M, Flyvbjerg A, Dahan R, Nephesh I, Safra T, Shiloni E, Raz I. Expression of the insulin-like growth factors and their receptors in adenocarcinoma of the colon [J]. Gut, 1999, 44(5): 704-708.doi:10.1136/gut.44.5.704.
[19] 宋姗姗. 水貂IGF-1R基因克隆、表达及多态性分析 [D]. 北京:中国农业科学院, 2016.
Song S S. Cloning, expression and polymorphism analysis of IGF-1R gene in mink [D]. Beijing:Chinese Academy of Agricultural Sciences, 2016.
[20] Duan J, Wainwright M S, Comeron J M, Saitou N, Sanders A R, Gelernter J, Gejman P V. Synonymous mutations in the human dopamine receptor D2(DRD2)affect mRNA stability and synthesis of the receptor [J]. Human Molecular Genetics, 2003, 12(3): 205-216. doi: 10. 1093/hmg/ddg055.