棉花是我国重要的纤维作物及经济作物,在国民经济中具有举足轻重的战略地位。目前,我国棉花在向新疆转移的大形势下,早熟作为育种的重要目标性状显得尤为重要[1]。适时开花是早熟棉培育的一个重要指标,也是棉花能否获得丰产的重要农艺性状之一[2]。分析花发育相关基因,对棉花品种熟期改良及实现棉花机械化采收具有重要意义。
磷脂酰乙醇胺结合蛋白(Phosphatidylethanolamine binding protein,PEBP)在自然界广泛分布。在动物方面的研究表明,PEBP蛋白参与MAPK/ERK信号通路调节细胞分化、迁移和黏附,参与NF-κB 信号通路调节细胞凋亡,参与GPCR 信号通路调控G 蛋白的信号转导等[3-5]。植物中PEBP蛋白主要参与开花调控通路、植株形态构建和ABA诱导的种子萌发信号等途径[6-8]。植物中TFL1(Terminal flower1)是磷脂酰乙醇胺结合蛋白中的1个亚组,该亚组基因的功能大部分集中在开花调控通路。拟南芥中TFL1通过维持茎顶端分生组织和花序分生组织的无限性,延长植株的营养生长过程,从而延迟植株的生殖生长进程[9]。Conti等[10]研究表明,TFL1蛋白能通过运输到达顶芽,并且调控拟南芥的植株结构,TFL1蛋白还能抑制AP1和LFY基因的表达并且保持花序的无限生长。对TFL1基因启动子结构的研究发现,该基因通过在营养器官的分生组织中表达来控制开花时间[11]。Liu等[12]在山茱萸中发现,ClTFL1过表达转化拟南芥后,拟南芥开花时间推迟,植株高度增加,顶端花芽和次生花序分枝增多。水稻中TFL1同源蛋白质RCN与Hd3a竞争是通过与14-3-3蛋白结合调控花发育[13]。棉花中GhTFL1基因与果枝形成发育相关[14-15]。
本研究依据棉花基因组测序结果[16-17],以棉花品种中棉所36为材料,克隆GhTFL1b基因,进行组织特异性、激素和胁迫处理,分析其表达模式,对GhTFL1亚组与GhFD进行互作分析,旨在摸清GhTFL1b表达调控的分子机制,从而为棉花早熟分子改良提供理论依据。
1 材料和方法
1.1 试验材料
试验所用棉花材料包括栽培种和半野生种,栽培种为中棉所36,半野生种为尖斑棉。DNA聚合酶、反转录酶、pMD18-T载体、质粒提取试剂盒、胶回收试剂盒和荧光定量试剂盒均购自TaKaRa公司;RNA提取试剂盒购自天根公司;大肠杆菌感受态DH5α购自康为世纪公司;pBI121载体及根癌农杆菌LBA4404由河南科技学院棉花基因工程实验室制备并保存。
1.2 材料处理及取样
用于光照试验的材料种植于中国农业科学院棉花研究所早熟组人工气候培养室,培养条件为长日照。二叶展平时期,短日照处理7 d,并将处理过后的棉花分2批继续培养,1批为长日照,光照/黑暗为14 h/10 h,光照时间为8:00-22:00;另一批为短日照,光照/黑暗为10 h/14 h,光照时间为8:00-18:00,温度均为28 ℃,子叶展平期到五叶展平期取样,取样后快速放入液氮中冷冻,并于-80 ℃冰箱中保存备用。利用棉花水培法来研究GhTFL1b在激素(赤霉素、脱落酸和水杨酸)和盐胁迫(NaCl)处理下的应答模式。所用材料为中棉所36水培苗,室内种植条件为14 h光照/10 h黑暗,温度为26 ℃。于三叶期水培液中增加激素和胁迫处理,以清水为对照。所用水杨酸(Salicylic acid,SA)浓度为200 μmol/L,脱落酸(Abscisic acid,ABA)和赤霉素(Gibberellins,GA)浓度为100 μmol/L,分别于处理后1,3,7,12,24 h进行根部取样,选取10株左右水培苗剪下幼嫩根部,吸取根部水分后液氮冷冻,并于-80 ℃冰箱中保存备用,试验重复3次,利用GraphPad Prism 5软件进行作图及统计分析。
1.3 生物信息学分析
研究中所用到的氨基酸序列下载自NCBI网站,使用ClustalX2软件进行多重序列比对,使用MEGA 6.06最大似然方法构建进化树。利用Compute pI/Mw(http://web.expasy.org/compute_pi/)对蛋白质理化性质进行预测和分析,并用CDD(https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml)数据库对其结构域进行预测。采用PlantCARE(http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)预测启动子区域顺式作用元件。
1.4 基因定量表达分析
使用天根多糖多酚RNA提取试剂盒提取RNA,所使用RNA的质量必须保证OD260/280为1.8~2.1,OD260/230大于1.8。使用TaKaRa RR047A试剂盒进行反转录,使用gDNA Eraser消化总RNA中残留的基因组DNA,进行15 min的反转录,合成cDNA。使用SYBR Green分析法进行荧光定量,所用试剂盒为TaKaRa RR420A,所用仪器为Applied Biosystems Q6实时荧光定量PCR仪。棉花中以ACTIN为内参,所用引物见表1。
1.5 重组载体构建和酵母转化
载体pGBKT7、pGADT7以及菌株Y2H和 Y187购自Clontech。以cDNA为模板,通过特异性Infusion引物扩增FD和GhTFL1亚组基因在棉花中的CDS,所用引物见表1。利用限制性内切酶EcoRⅠ和SacⅠ双酶切载体pGADT7,利用EcoRⅠ和PstⅠ双酶切载体pGBKT7,形成黏性末端,利用Infusion连接试剂盒进行连接。将连接后的新鲜菌液送金唯智生物科技有限公司测序,并保存测序正确的单克隆菌液以供后续试验使用。参考Zhang等[18]的方法进行酵母转化和酵母双杂交试验。
表1 棉花GhTFL1b基因克隆及 表达所用引物(GhTFL1a、GhTFL1c、GhTFL1d)
Tab.1 Primers used in GhTFL1b gene cloning and expression in upland cotton
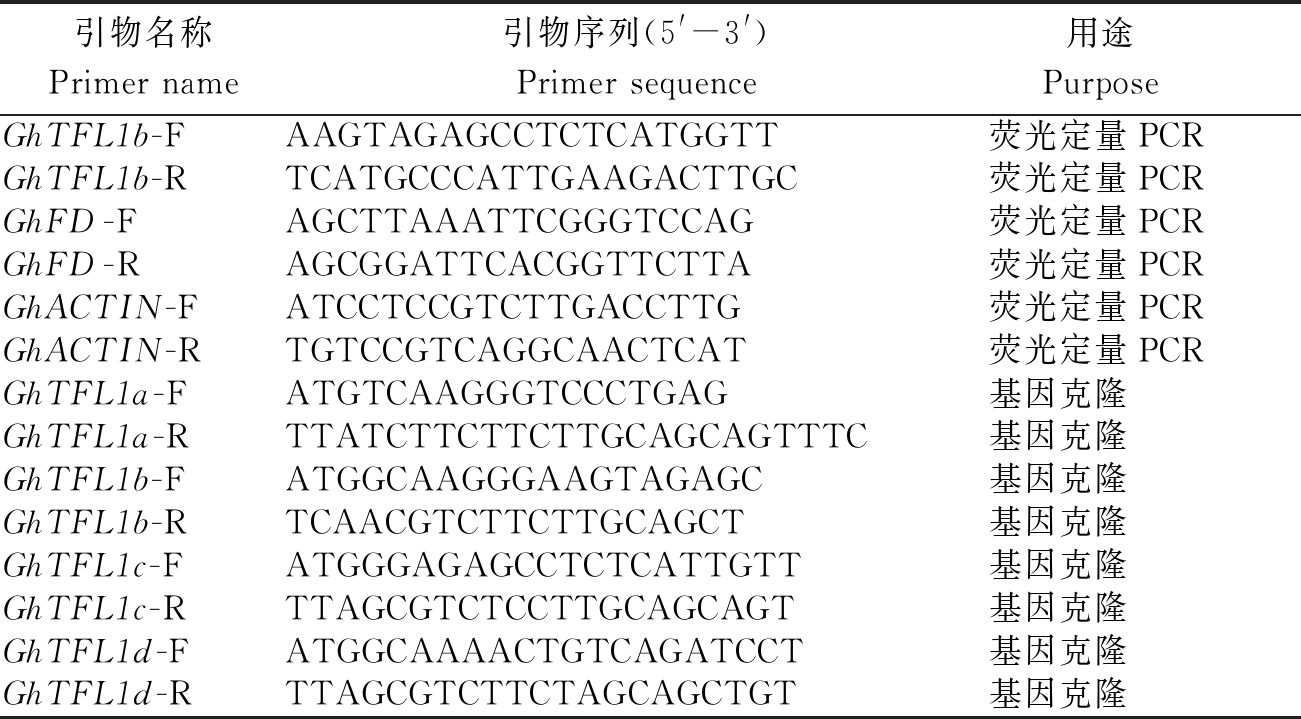
引物名称Primer name引物序列(5'-3')Primer sequence 用途PurposeGhTFL1b-FAAGTAGAGCCTCTCATGGTT荧光定量PCRGhTFL1b-RTCATGCCCATTGAAGACTTGC荧光定量PCRGhFD-FAGCTTAAATTCGGGTCCAG荧光定量PCRGhFD-RAGCGGATTCACGGTTCTTA荧光定量PCRGhACTIN-FATCCTCCGTCTTGACCTTG荧光定量PCRGhACTIN-RTGTCCGTCAGGCAACTCAT荧光定量PCRGhTFL1a-FATGTCAAGGGTCCCTGAG基因克隆GhTFL1a-RTTATCTTCTTCTTGCAGCAGTTTC基因克隆GhTFL1b-FATGGCAAGGGAAGTAGAGC基因克隆GhTFL1b-RTCAACGTCTTCTTGCAGCT基因克隆GhTFL1c-FATGGGAGAGCCTCTCATTGTT基因克隆GhTFL1c-RTTAGCGTCTCCTTGCAGCAGT基因克隆GhTFL1d-FATGGCAAAACTGTCAGATCCT基因克隆GhTFL1d-RTTAGCGTCTTCTAGCAGCTGT基因克隆
2 结果与分析
2.1 棉花GhTFL1b基因的序列分析
棉花GhTFL1亚组中有4个基因,分别是GhTFL1a、GhTFL1b、GhTFL1c和GhTFL1d。GhTFL1b基因位于棉花A亚组第11号染色体上,编码框长519 bp,编码172氨基酸。经理化性质预测可知,其编码蛋白质分子质量约为19.6 ku,等电点为9.41。进化树分析结果表明,棉花GhTFL1b与拟南芥AtTFL1亲缘关系相近(图1-A)。根据TAIR网站(http://www.arabidopsis.org/)提供的拟南芥TFL1氨基酸序列进行序列比对,该氨基酸序列具有比较保守的磷脂酰乙醇胺结合蛋白结构域(图1-B)。

A.进化树分析;B.氨基酸多重序列比对;*代表氨基酸完全一致,AtBFT、AtTFL1、AtATC序列号分别为NM_125597、U77674、NM_128315。
A.Phylogenetic analysis of GhTFL1 and their homologous genes in Arabidopsis; B.Alignment of amino acid sequences of GhTFL1b and their homologous genes in Arabidopsis; *indicated the same amino acid,the ID of AtBFT,AtTFL1 and AtATC is NM_125597,U77674 and NM_128315,respectively.
图1 棉花GhTFL1b进化树及氨基酸多重序列比对
Fig.1 Phylogenetic and amino acid multiple sequence alignment analysis of GhTFL1b in cotton
2.2 GhTFL1b基因的表达分析
对GhTFL1b基因进行组织表达特异性分析表明,该基因在花中表达量最高,其次是顶芽和根(图2)。为研究GhTFL1b是否受光周期影响,利用光周期试验处理半野生种和栽培种材料,对不同时期幼苗叶片取样分析(图3)。结果表明,GhTFL1b在半
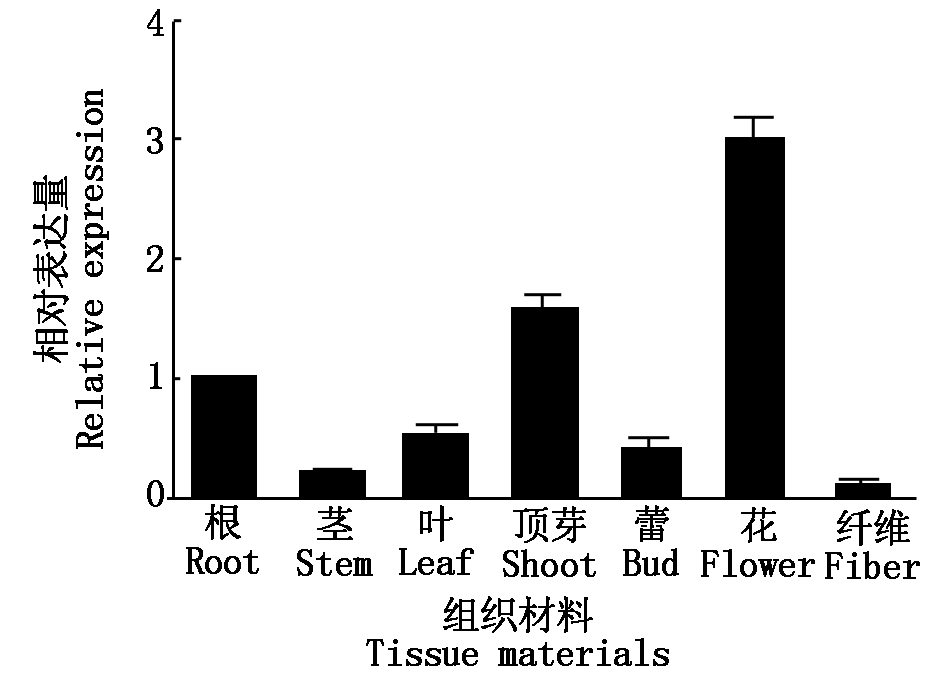
图2 棉花GhTFL1b组织特异性表达
Fig.2 Organ-specific expression of GhTFL1b in cotton
野生种和栽培种中表达量无明显变化趋势。该结果说明,GhTFL1b基因在花中优势表达,而且从半野生种到栽培种的进化过程中,GhTFL1b对不同光照条件变化趋势不敏感。
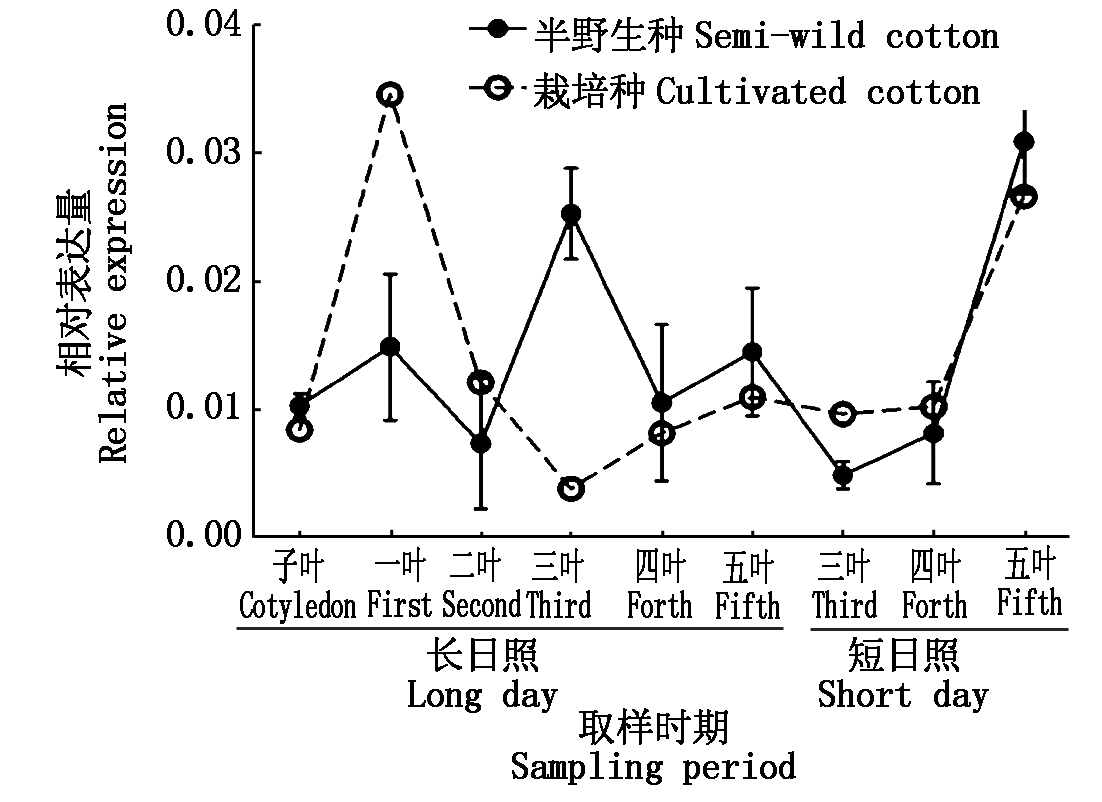
图3 棉花GhTFL1b在栽培种和半野生种中表达模式
Fig.3 Expression patterns of GhTFL1b genes in cultivated cotton and semi-wild cotton
2.3 GhTFL1b启动子分析及其对外源激素的应答
GhTFL1b基因拥有磷脂酰乙醇胺结合蛋白的保守结构域,因此分析启动子的顺式作用元件对于揭示GhTFL1b的功能非常必要。从中棉所棉花基因组数据库中获取GhTFL1b基因起始密码子上游2 000 bp的序列,并利用PlantCARE数据库对其顺式作用元件进行预测。结果如表2所示,该基因的启动子主要存在2大类顺式作用元件,一类是光响应元件和生物钟节律响应元件,一类是胁迫响应元件,如与胁迫相关的茉莉酸、脱落酸和干旱胁迫等元件,表明GhTFL1b的表达可能受光、生物钟、逆境及植物激素等的调控。
表2 棉花GhTFL1b启动子顺式作用元件预测
Tab.2 The predicted cis-acting elements of GhTFL1b promoter in cotton
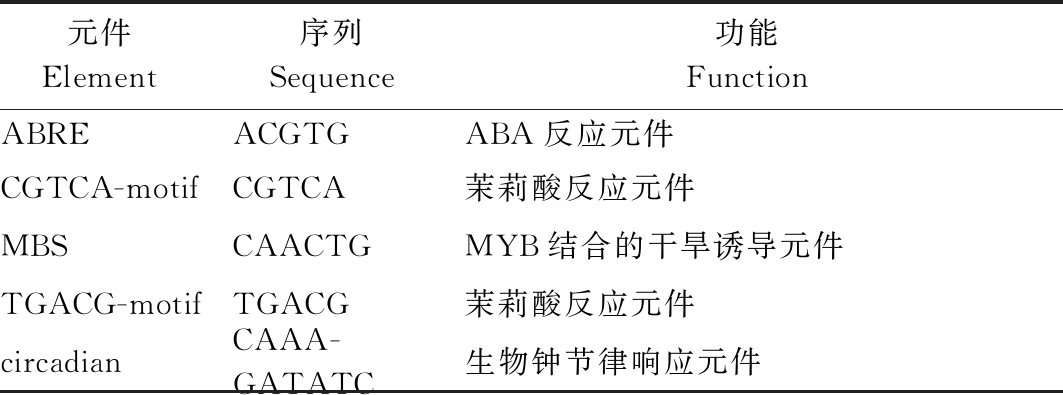
元件 Element 序列 Sequence 功能 Function ABREACGTGABA反应元件CGTCA-motifCGTCA茉莉酸反应元件MBSCAACTGMYB结合的干旱诱导元件TGACG-motifTGACG茉莉酸反应元件circadianCAAA-GATATC生物钟节律响应元件
胁迫响应24 h内GhTFL1b表达量变化如图4。经SA处理3 h后GhTFL1b的表达量开始下降,7 h降到最低值(图4-A)。经ABA处理(图4-B)1 h后GhTFL1b表达量开始下降,到3 h达到最小值,随后开始上升。经GA处理后(图4-C),GhTFL1b的表达量与对照相比,在3 h达到峰值,随后开始下降。盐胁迫后GhTFL1b表达量从3 h开始持续上升,到24 h到达最高,表达量约是对照组的6.4倍(图4-D)。以上结果表明,SA能抑制棉花GhTFL1b的表达,盐胁迫能促进GhTFL1b的表达,而ABA和GA对GhTFL1b的转录无明显作用,GhTFL1b可以响应SA和盐胁迫。
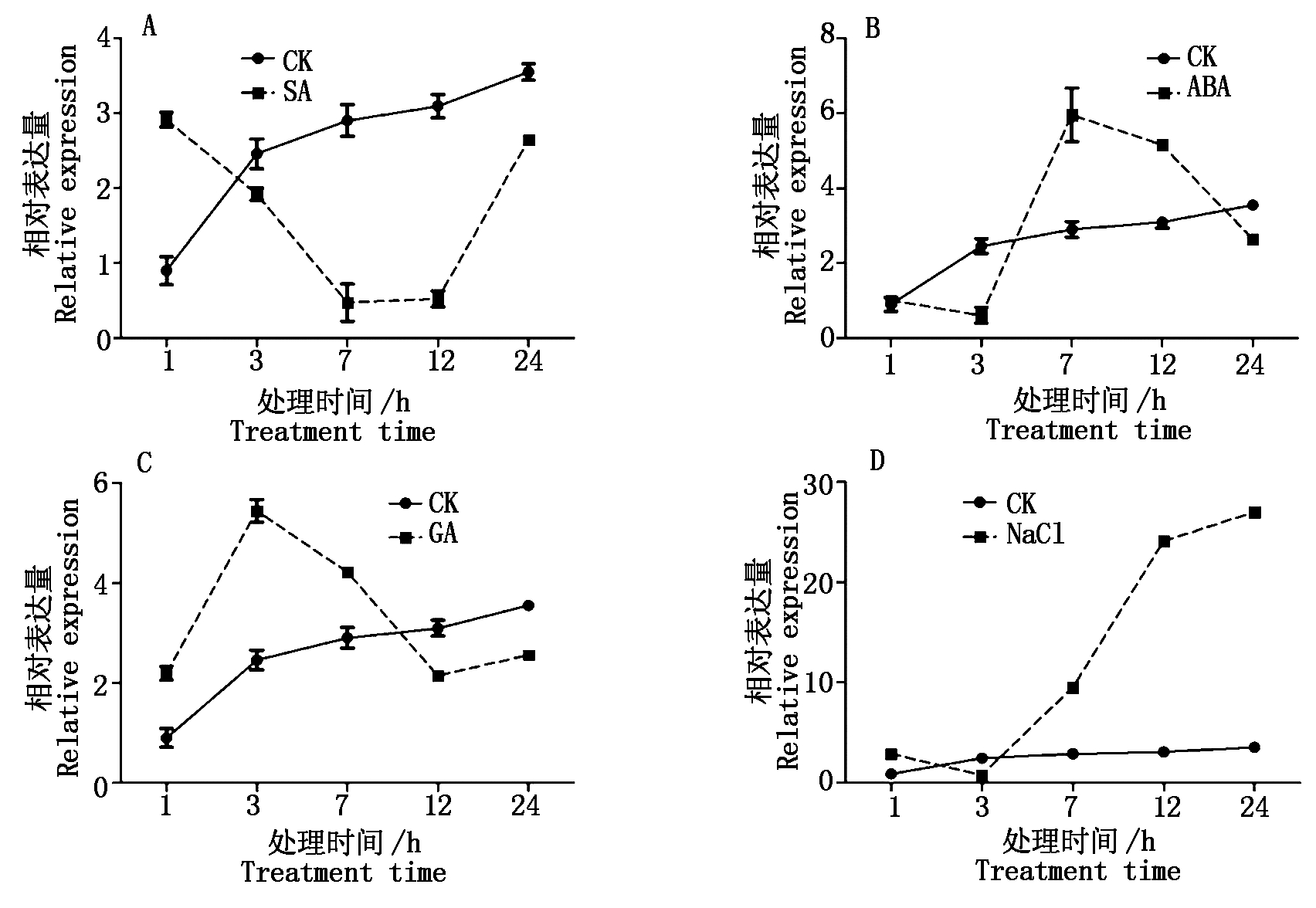
图4 棉花水培苗经过处理后24 h内GhTFL1b基因表达量
Fig.4 GhTFL1b expression profiles in the first 24 hours after the cotton seeding were treated using hydroponics
2.4 GhTFL1亚组与GhFD蛋白的互作
GhFD是一个b-ZIP转录因子[18],该基因在茎端分生组织中优势表达(图5-A)。克隆棉花GhFD基因,连接到pGBKT7载体,检测是否有自我激活活性和毒性。结果表明,该蛋白质无毒性,但是存在自我激活活性。将GhFD连接到pGADT7载体,将GhTFL1a/GhTFL1b/GhTFL1c/GhTFL1d蛋白分别连接到pGBKT7载体。双菌株Y187和Y2H混合培养结果显示,GhFD蛋白与GhTFL1b存在互作(图5-B)。
3 结论与讨论
开花是棉花的重要农艺性状之一。花期的早晚影响产量的高低,也影响品种的区域适应性。随着棉花基因组的测序完成[16-17],对棉花功能基因进行的相关研究越来越多[19-21],而针对棉花开花基因启动子的分析和蛋白质表达调控等的研究相对较少。从棉花基因组数据库中共检索到4条TFL1亚组基因,本研究克隆了GhTFL1b,组织表达和蛋白质调控分析表明,GhTFL1b可以响应水杨酸和盐胁迫,该基因编码的蛋白质可与转录因子GhFD蛋白进行互作。
水杨酸是与植物胁迫相关的重要激素,也是遮荫条件下改善植物光合作用并延长花期的重要因子[22],其不仅能依赖CO基因调控的光周期途径调节植物花期,也可以通过抑制开花负调控因子FLC在自主途径中促进开花[23-24]。盐胁迫是常见的一种逆境条件,会对植物根系细胞结构产生较大影响,如质膜透性增大,脂质过氧化作用增强,脯氨酸含量增加等[25-26]。拟南芥TFL1家族中BFT基因在高盐胁迫下通过介导FT-FD模型提供合适的保护机制,适应光周期途径并促使开花[27-28]。本研究结果表明,盐胁迫处理后GhTFL1b基因高调表达,水杨酸诱导可使其表达受到抑制,光周期变化不影响该基因表达模式。
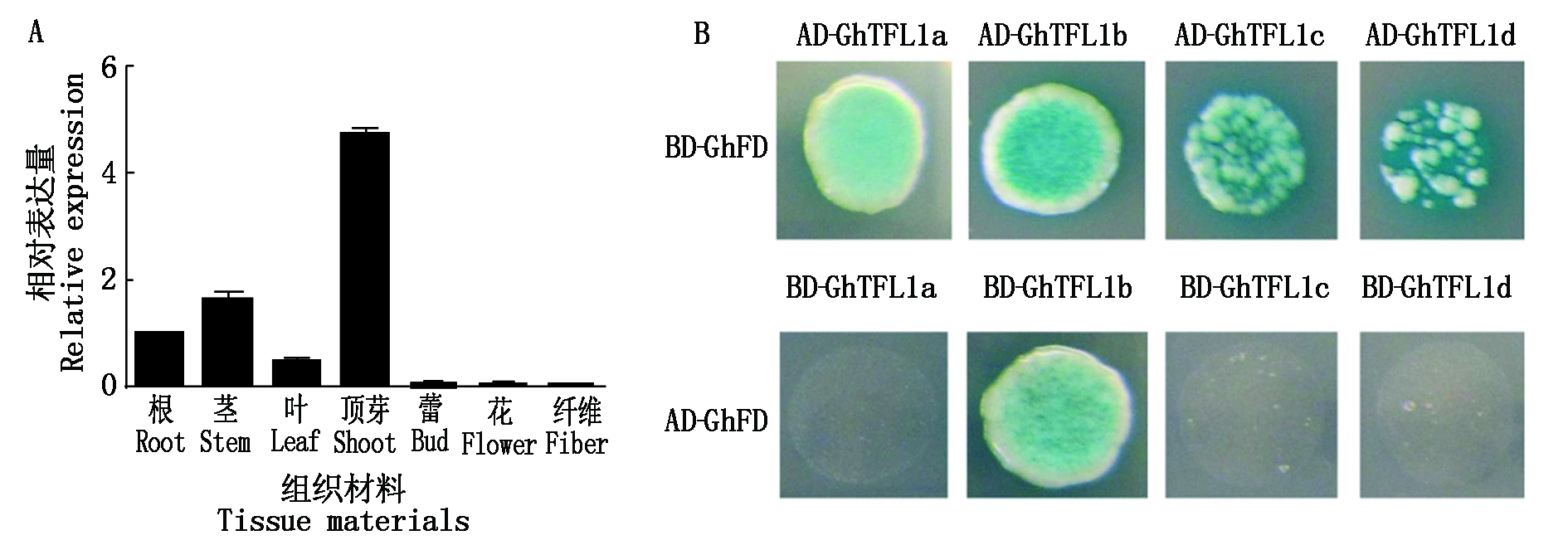
AD.转录激活结构域;BD.DNA结合结构域。
AD.Activation domain; BD.DNA binding domain.
图5 棉花GhFD基因组织表达(A)及GhFD与GhTFL1a、GhTFL1b、GhTFL1c和GhTFL1d蛋白质互作分析(B)
Fig.5 Analysis of organ-specific expression of GhFD in cotton and protein interaction of GhFD with GhTFL1a,GhTFL1b,GhTFL1c and GhTFL1d
FD属于b-ZIP转录因子,拟南芥FD蛋白能结合FT和TFL1调控开花时间和花的发育[29-30]。棉花基因组有10个GhFD基因,其中5条来自A组,5条来自D组[31]。Prewitt等[15]研究证实其中1个棉花FD蛋白与GhSP、GhTFL1、GhBFT蛋白互作。本研究结果也表明,棉花中1个GhFD能与GhTFL1b互作。棉花其余几个FD蛋白与磷脂酰乙醇胺结合蛋白家族间的关系还有待进一步研究。
棉花GhTFL1b与拟南芥TFL1亲缘关系相近。拟南芥TFL1基因能延迟开花时间[30],黄瓜方面的研究表明CsTFL1b能推迟开花并影响花序形态发育[32]。马铃薯StCEN能调节茎尖分生组织的发育,进一步调控株型发育[33]。因此,还需通过转基因棉花验证,并对转基因后代株系进行胁迫处理,进一步研究掌握GhTFL1b基因的功能。
[1] 董承光,王娟,周小凤,马晓梅,李生秀,王旭文,肖光顺,李保成. 新疆早熟棉花早熟性状的遗传分析[J].西北农业学报,2014,23(12):96-101. doi:10.7606/j.issn.1004-1389.
Dong C G,Wang J,Zhou X F,Ma X M,Li S X,Wang X W,Xiao G S,Li B C. Inheritance of earliness traits in xinjiang early-maturity upland cotton(G. hirsutum L.)[J]. Acta Agriculture Boreali-occident Sinica,2014,13(12):96-101.
[2] 喻树迅,王寒涛,魏恒玲,宿俊吉. 棉花早熟性研究进展及其应用[J].棉花学报,2017,29(S1):1-10. doi:10.11963/1002-7807.
Yu S X,Wang H T,Wei H L,Su J J. Research progress and application of early maturity in upland cotton[J]. Cotton Science,2017,29(S1):1-10.
[3] Okita K,Matsukawa N,Maki M,Nakazawa H,Katada E,Hattori M,Akatsu H,Borlongan C V,Ojika K. Analysis of DNA variations in promoter region of HCNP gene with Alzheimer′s disease[J].Biochemical and Biophysical Research Communications,2009,379(2):272-276. doi:10.1016/j.bbrc.2008.12.037.
[4] Uematsu N,Matsukawa N,Kanamori T,Arai Y,Sagisaka T,Toyoda T,Yoshida M,Ojika K. Overexpression of hippocampal cholinergic neurostimulating peptide in heterozygous transgenic mice increases the amount ofChAT in the medial septal nucleus[J]. Brain Research,2009,1305:150-157. doi:10.1016/j.brainres.2009.09.112.
[5] Morecroft I,Doyle B,Nilsen M,Kolch W,Mair K,Maclean M R. Mice lacking the Raf 1 kinase inhibitor protein exhibit exaggerated hypoxia induced pulmonary hypertension[J].British Journal of Pharmacology,2011,163(5):948-963. doi:10.1111/j.1476-5381.2011.01305.x.
[6] Tamaki S,Matsuo S,Wong H L,Yokoi S,Shimamoto K. Hd3a protein is a mobile flowering signal in rice[J]. Science,2007,316(5827):1033-1036. doi:10.1126/science.1141753.
[7] Baumann K,Venail J,Berbel A,Domenech M J,Money T,Conti L,Hanzawa Y,Madueno F,Bradley D. Changing the spatial pattern of TFL1 expression reveals its key role in the shoot meristem in controlling Arabidopsis flowering architecture[J]. Journal of Experimental Botany,2015,66(15):4769-4780. doi:10.1093/jxb/erv247.
[8] Xi W,Liu C,Hou X,Yu H. Mother of FT and TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis[J]. Plant Cell,2010,22(6):1733-1748. doi:10.1105/tpc.109.073072.
[9] Hanzawa Y,Money T,Bradley D. A single amino acid converts a repressor to an activator of flowering[J]. Proceeding of the National Academy of Science of the United States of America,2005,102(21):7748-7753. doi:10.1073/pnas.0500932102.
[10] Conti L,Bradley D. TERMINAL FLOWER1 is a mobile signal controlling Arabidopsis architecture[J]. Plant Cell,2007,19(3):767-778. doi:10.1105/tpc.106.049767.
[11] Serrano Mislata A,Fernandez Nohales P,Domenech M J,Hanzawa Y,Bradley D,Madueno F. Separate elements of the TERMINAL FLOWER 1 cis-regulatory region integrate pathways to control flowering time and shoot meristem identity[J]. Development,2016,143(18):3315-3327. doi:10.1242/dev.135269.
[12] Liu X,Zhang J,Abuahmad A,Franks R G,Xie D Y,Xiang Q Y. Analysis of two TFL1 homologs of dogwood species(Cornus L.)indicates functional conservation in control of transition to flowering[J]. Planta,2016,243(5):1129-1141. doi:10.1007/s00425-016-2466-x.
[13] Kaneko-Suzuki M,Kurihara-Ishikawa R,Okushita-Terakawa C,Kojima C,Nagano-Fujiwara M,Ohki I,Tsuji H,Shimamoto K,Taoka K I. TFL1-Like proteins in rice antagonize rice FT-like protein in inflorescence development by competition for complex formation with 14-3-3 and FD[J]. Plant Cell Physiology,2018,59(3):458-468. doi:10.1093/pcp/pcy021.
[14] Si Z F,Liu H,Zhu J K,Chen J D,Wang Q,Fang L,Gao F K,Tian Y,Chen Y L,Chang L J,Liu B L,Han Z G,Zhou B L,Hu Y,Huang X Z,Zhang T Z. Mutation of SELF-PRUNING homologs in cotton promotes short-branching plant architecture[J].Journal of Experimental Botany,2018,69(10):2543-2553. doi:10.1093/jxb/ery093.
[15] Prewitt S F,Ayre B G,McGarry R C. Cotton CENTRORADIALIS/TERMINAL FLOWER 1/SELF PRUNING genes functionally diverge to differentially impact architecture[J]. Journal ofExperimental Botany,2018,69(22):5403-5417.doi:10.1093/jxb/ery324.
[16] Li F G,Fan G Y,Lu C R,Xiao G H,Zou C S,Kohel R J,Ma Z Y,Shang H H,Ma X F,Wu J Y,Liang X M,Huang G,Percy R G,Liu K,Yang W H,Chen W B,Du X M,Shi C C,Yuan Y L,Ye W W,Liu X,Zhang X Y,Liu W Q,Wei H L,Wei S J,Huang G D,Zhang X L,Zhu S J,Zhang H,Sun F M,Wang X F,Liang J,Wang J H,He Q,Huang L H,Wang J,Cui J J,Song G L,Wang K B,Xu X,Yu J Z,Zhu Y X,Yu S X. Genome sequence of cultivated upland cotton(Gossypium hirsutum TM 1)provides insights into genome evolution[J]. Nature Biotechnology,2015,33(5):524-530. doi:10.1038/nbt.3208.
[17] Zhang T Z,Hu Y,Jiang W K,Fang L,Guan X Y,Chen J D,Zhang J B,Saski C A,Scheffler B E,Stelly D M,Hulse Kemp A M,Wan Q,Liu B L,Liu C X,Wang S,Pan M Q,Wang Y K,Wang D W,Ye W X,Chang L J,Zhang W P,Song Q,Kirkbride R C,Chen X Y,Dennis E,Llewellyn D J,Peterson D G,Thaxton P,Jones D C,Wang Q,Xu X Y,Zhang H,Wu H T,Zhou L,Mei G F,Chen S Q,Tian Y,Xiang D,Li X H,Ding J,Zuo Q Y,Tao L N,Liu Y C,Li J,Lin Y,Hui Y Y,Cao Z S,Cai C P,Zhu X F,Jiang Z,Zhou B L,Guo W Z,Li R Q,Chen Z J. Sequencing of allotetraploid cotton(Gossypium hirsutum L. acc. TM 1)provides a resource for fiber improvement[J]. Nature Biotechnology,2015,33(5):531-537. doi:10.1038/nbt.3207.
[18] Zhang X H,Wang C C,Pang C Y,Wei H L,Wang H T,Song M Z,Fan S L,Yu S S. Characterization and functional analysis of PEBP family genes in upland cotton(Gossypium hirsutum L.)[J].PLoS One,2016,11(8):e0161080. doi:10.1371/journal.pone.0161080.
[19] 石建斌,王宁,周红,许庆华,乔文青,严根土. 陆地棉中赤霉素合成途径关键酶基因的时空表达变化[J].华北农学报,2018,33(04):9-16.doi:10.7668/hbnxb.2018.04.002.
Shi J B,Wang N,Zhou H,Xu Q H,Qiao W Q,Yan G T. The temporal spatial expression of gibberellin key metabolic enzyme in upland cotton[J]. Acta Agriculturae Boreali-Sinica,2018,33(4):9-16.
[20] 王聪聪,张晓红,王小艳,张盼,范术丽,庞朝友,马启峰,魏恒玲,王寒涛,宿俊吉,喻树迅. 棉花开花相关基因GhFLP5的表达及功能分析[J].中国农业科学,2017,50(12):2220-2231. doi:10.3864/j.issn.0578-1752.2017.12.003.
Wang C C,Zhang X H,Wang X Y,Zhang P,Fan S L,Pang C Y,Ma Q F,Wei H L,Wang H T,Su J J,Yu S X. The expression patterns and function analysis of GhFLP5,a gene related to flowering in upland cotton(Gossypium hirsutum L.)[J]. Scientia Agricultura Sinica,2017,50(12):2220-2231.
[21] 张文香,庞朝友,范术丽,宋美珍,魏恒玲,喻树迅. 棉花SVP-like基因GhMADS29的克隆与表达分析[J].安徽农业科学,2015,43(15):28-31. doi:10.13989/j.cnki.0517-6611.2015.15.012.
Zhang W X,Pang C Y,Fan S L,Song M Z,Wei H L,Yu S X. Molecular cloning and expression analysis of SVP-like gene GhMADS29 from Gossypium hirsutum L.[J].Journal of Anhui Agricultural Science,2015,43(15):28-31.
[22] 艾星梅,黄美娟,黄海泉. 连续弱光下水杨酸对重瓣百合Elena开花前后光合特性及生长的影响[J].河南农业科学,2018,47(4):93-98. doi:10.15933/j.cnki.1004-3268.2018.04.017.
Ai X M,Huang M J,Huang H Q. Effects of salicylic acid on photosynthetic characteristics and growth of double lily elena before and after flowering under continuous weak light conditions[J].Journal of Henan Agricultural Sciences,2018,47(4):93-98.
[23] Wada K C,Yamada M,Shiraya T,Takeno K. Salicylic acid and the flowering gene FLOWERING LOCUST homolog are involved in poor-nutrition stress-induced flowering of Pharbitis nil[J].Journal of Plant Physiology,2010,167(6):447-452. doi:10.1016/j.jplph.2009.10.006.
[24] Martinez C,Pons E,Prats G,Leon J. Salicylic acid regulates flowering time and links defence responses and reproductive development[J]. Plant Journal,2004,37(2):209-217. doi:10.1046/j.1365-313X.2003.01954.x.
[25] 姜艳丽,史华平,杨艳兵,尹晓斐,王计平. NaCl胁迫对棉花叶片及根系超微结构的影响[J].华北农学报,2014,29(3):95-100.doi:10.7668/hbnxb.2014.03.018.
Jiang Y L,Shi H P,Yang Y B,Yin X F,Wang J P. Effect of NaCl stress on ultrastructure of mesophyll cells and root cells in cotton[J].Acta Agriculturae Boreali-Sinica,2014,29(3):95-100.
[26] 胡根海,张晓红,付远志,董娜,王清连. 棉花过表达超氧化物歧化酶基因对植株耐盐性的影响[J].华北农学报,2017,32(6):54-59. doi:10.7668/hbnxb.2017.06.008.
Hu G H,Zhang X H,Fu Y Z,Dong N,Wang Q L. Effects of overexpression of cotton superoxide dismutase geneson salt tolerant capability in upland cotton[J].Acta Agriculturae Boreali-Sinica,2017,32(6):54-59.
[27] Ryu J Y,Lee H J,Seo P J,Jung J H,Ahn J H,Park C M. The Arabidopsis floral repressor BFT delays flowering by competing with FT for FD binding under high salinity[J].Molelular Plant,2014,7(2):377-387. doi:10.1093/mp/sst114.
[28] Ryu J Y,Park C M,Seo P J. The floral repressor BROTHER OF FT AND TFL1(BFT)modulates flowering initiation under high salinity in Arabidopsis[J]. Molecular Cells,2011,32(3):295-303. doi:10.1007/s10059-011-0112-9.
[29] Abe M,Kobayashi Y,Yamamoto S,Daimon Y,Yamaguchi A,Ikeda Y,Ichinoki H,Notaguchi M,Goto K,Araki T. FD,a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex[J]. Science,2005,309(5737):1052-1056. doi:10.1126/science.1115983.
[30] Hanano S,Goto K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression[J]. Plant Cell,2011,23(9):3172-3184. doi:10.1105/tpc.111.088641.
[31] 张彦楠,于秀立,吕新华,刘慧,黄先忠. 棉花bZIP转录因子FD基因的发掘和GhFD基因的组织表达分析[J].分子植物育种,2016,14(9):2250-2260. doi:10.13271/j.mpb.014.002250.
Zhang Y N,Yu X L,L X H,Liu H,Huang X Z. Genome-wide identification of bZIPtranscription factor FD genes and expression patterns analysis of GhFD genes in cotton[J].Molecular Plant Breeding,2016,14(9):2250-2260.
[32] Zhao W,Gu R,Che G,Cheng Z,Zhang X.CsTFL1b may regulate the flowering time and inflorescence architecture in cucumber(Cucumis sativus L.)[J].Biochemical and Biophysical Research Communications,2018,499(2):307-313. doi:10.1016/j.bbrc.2018.03.153.
[33] Morris W L,Alamar M C,Lopez Cobollo R M,Castillo J C,Bennett M,Van der Kaay J,Stevens J,Kumar Sharma S,McLean K,Thompson A J,Terry L A,Turnbull C G N,Bryan G J,Taylor M A. A member of the TERMINALFLOWER1/CENTRORADIALIS gene family controls sprout growth in potato tubers[J]. Journal of Experimental Botany,2018,70(3):835 843. doi:10.1093/jxb/ery387.