START(The lipid/sterol-binding StAR-related lipid transfer protein domains)结构域广泛存在于动物、植物和微生物中,此结构域调控着磷脂运输和脂质代谢。在人类和动物很多蛋白质中含有START结构域,与疾病的发生密切相关;在植物中START结构域往往和其他植物特有的结构域同时存在于一个蛋白质中,调控着植物各个生长发育过程,如ATML1(A. thaliana MERISTEM LAYER1)在皮层的发育过程中起重要的调控作用[1],PDF2(PROTODERMAL FACTOR2)可调控花器官的形成[2],GL2(GLABRA2)调控表皮毛的生长[3]。ATML1、PDF2和GL2均属于START结构域蛋白家族中的HD-ZLZ START(Homeodomain zipper-loop-zipper StAR-related lipid transfer)亚家族,这类亚家族成员属于转录因子,START结构域在这些转录因子中的作用类似于动物中的甾醇类激素受体,通过结合脂类或甾醇类物质来调控转录过程。拟南芥中含START结构域家族成员共35个,但已知功能的仅有9个,均为转录因子家族[4]。在START结构域家族中,还有PH-START(Pleckstrin homology StAR-related lipid transfer)亚家族,但此亚家族成员在调控植物生长发育方面的功能还未见报道。
拟南芥中PH-START蛋白亚家族由At4g19040、At5g45560、At3g54800、At2g28320 4个成员组成。除有研究发现At4g19040突变体可增强拟南芥对白粉菌(Erysiphe cichoracearum)的抗性外[5],此亚家族成员对拟南芥生长发育的调控作用未见报道。PH(Pleckstrin homology)结构域首先在普雷克底物蛋白中识别,大约含有120个氨基酸残基,是血小板中蛋白激酶C(PKC)的主要底物[6-7]。PH结构域是很多与细胞膜结构结合的第二信使蛋白质所具有的结构域。已证实PH结构域参与了细胞内信号传导、细胞骨架组成、膜磷脂的转运和修饰[8-9]。在人类基因组中,已经发现了252 种蛋白质含有不同结构的PH结构域[10],在酿酒酵母中,有33种蛋白质具有不同结构的PH结构域[11]。迄今为止,一些PH结构域的结构已经通过核磁共振和X射线获得,尽管不同PH结构域之间的序列相似性很低,但其三维结构却具有显著的保守性[12]。虽然已经在不同基因组中识别了大量的PH结构域,但PH结构域的功能尚不清楚[13]。PH结构域首次被识别时,认为是一个蛋白质结合域,并且可以识别蛋白质配体,例如异源三聚体G蛋白的β/γ亚基、WD40重复蛋白和酪氨酸激酶等[14]。然而,PH结构域最显著的功能是它们与磷脂(例如磷酸肌醇或肌醇磷脂酸)结合的能力[15],与磷酸肌醇类物质的结合可以使具有PH结构域的蛋白质对脂类细胞信使做出反应,从而转移至细胞膜结构上。蛋白磷脂酶C(PLC)家族具有PH结构域,PLC酶是存在于胞浆膜上的一个关键酶,可以水解磷脂酰肌醇4,5-二磷酸(PIP2),产生1,4,5-磷酸三肌醇和二酰甘油2个第二信号分子,调节细胞内的Ca2+释放和激活蛋白激酶C[15]。
本研究以PH-START亚家族成员At2g28320(命名为PH-START1)作为研究对象,分析其时空表达特性,探究其在拟南芥生长发育中的作用,为更好地促控植物生长发育、提高作物产量提供理论依据。
1 材料和方法
1.1 试验材料
野生型拟南芥(Arabidopsis thaliana):Columbia(Col-0),由河北农业大学植物生理和分子病理学重点实验室种植;拟南芥PH-START1(At2g28320)基因的T-DNA插入突变体(SALK-060171、SALK-120376),购自美国ABRC(Arabidopsis Biological Resource Center)突变体库。拟南芥生长培养条件为:温度22 ℃,光周期16 h光照/8 h黑暗。
1.2 试验方法
1.2.1 PH-START1基因表达量分析 为研究PH-START1基因在拟南芥的时空表达情况,以Col-0野生型为材料,分别以根、茎、不同位置的莲座叶、不同花期的花、授粉后不同时间的角果为材料,提取总RNA,经反转录后,利用Real-time PCR分析PH-START1基因的表达情况。
1.2.2 PH-START1过表达转基因拟南芥的获得 以PH-START1基因的CDS序列和拟南芥过表达载体pSN1301的MCS分析其酶切位点并选定为BamH Ⅰ和Kpn Ⅰ,以野生型拟南芥的cDNA作为模板,扩增PH-START1目的序列。对PCR扩增得到的目的片段经胶回收后与克隆载体pMD19-T Vector连接,转化大肠杆菌,对筛选得到的阳性克隆菌液检测并测序,将测序正确的菌液提取质粒,双酶切回收目的条带,然后与载体片段连接、转化、验证。将最终得到的含有目的基因的过表达载体命名为pSN1301-PH-START1。用电击法转化农杆菌GV3101,利用花絮侵染法转化野生型拟南芥,抗生素筛选阳性苗[16],并用Real-time PCR进行基因表达量的验证。
1.2.3 PH-START1 T-DNA插入突变体的获得 将购买的PH-START1基因突变体种子播种在含有Kan抗性的MS平板上(Kan:100 mg/L)筛选,选取叶片鲜绿下胚轴较长的植株移栽,收获种子继续筛选并进行验证。
三引物法:提取T3不再出现分离的突变体植株幼苗和野生型拟南芥的DNA,以T-DNA Primer Design(http://signal.salk.edu/tdnaprimers.2.html)设计插入位点上下游的引物LP、RP,以LBb1作为T-DNA特异引物,通过琼脂糖凝胶电泳验证T-DNA插入位点。
PH-START1基因表达量验证:提取28 d的Col-0野生型拟南芥和PH-START1突变体植株的RNA,反转录后用SqRT-PCR和Real-time PCR 验证PH-START1基因的表达情况。
1.2.4 拟南芥叶面积的测定 将生长环境完全一致的野生型和突变体植株相同部位的叶片展平,用Image J软件测量叶片的面积,野生型和突变体各测量30个叶片。
2 结果与分析
2.1 PH-START1时空表达分析
分别提取野生型拟南芥不同生长时期的根、茎、莲座叶、花、角果的总RNA并反转录,然后利用Real-time PCR分析PH-START1基因的表达量(图1)。结果发现,在拟南芥的各个器官中均有PH-START1基因的表达,在叶中表达最多,然后依次是根、花、角果和茎,且在各个器官中的表达差异达极显著水平(图1-A);从子叶期到幼苗期,根中PH-START1基因的表达量极显著上升,但到成苗期根中PH-START1的表达无显著改变(图1-B);对第2,4,6,8,10片莲座叶中PH-START1的表达分析发现,每片莲座叶中均有PH-START1的表达,在第6片莲座叶中的表达量最高,其次是第2片莲座叶,在第8,10片莲座叶中表达量最低(图1-C);授粉后的角果中PH-START1均有表达,授粉后第5天(5DAP)的角果中PH-START1表达量极显著高于3 d的角果,而在以后时期角果的表达量更低(图1-D);比较不同花期的花中PH-START1表达量,发现随着花的发育,花中PH-START1表达量逐渐增高,差异均达极显著水平(图1-E);比较花器官中花萼、花瓣、雄蕊、雌蕊PH-START1基因的表达情况,发现雄蕊中PH-START1的表达量最高(差异极显著)、其次是花瓣和萼片,而雌蕊中最少,差异达极显著水平(图1-F)。

不同小写和大写字母分别表示5%和1%水平差异显著。图9同。
Different small and capital letters indicate 5% and 1% significant level respectively.The same as Fig.9.
图1 拟南芥不同生长时期和不同组织部位中PH-START1基因的表达分析
Fig.1 The expression analysis of PH-START1 in different growth stages and different tissues in Arabidopsis
2.2 PH-START1过表达载体的构建
根据PH-START1 CDS序列引物扩增得到PH-START1 CDS序列,经琼脂糖凝胶电泳检测,在2 214 bp处得到单一条带,与PH-START1片段大小一致(图2-A)。回收目的条带,并与克隆载体pMD19-T vector 16 ℃连接1 h,转化E.coli DH5α,PCR检测阳性菌落(图2-B)。选择阳性克隆进行测序,并提取测序正确的阳性克隆质粒,用BamH Ⅰ和Kpn Ⅰ对其双酶切(图2-C),回收目的片段。目的片段与pSN1301载体片段用T4 DNA Ligase在16 ℃过夜连接,转化E.coli DH5α后对阳性克隆进行PCR检测(图2-D)。将阳性克隆用BamH Ⅰ和Kpn Ⅰ进行双酶切验证,得到PH-START1目的条带(图2-E),表明PH-START1过表达载体构建成功,命名为35S∶pSN1301-PH-START1。将构建好的35S∶pSN1301-PH-START1转化GV3101,用基因序列引物检测阳性克隆,得到PH-START1基因目的条带(图2-F),说明构建好的过表达载体成功转入农杆菌感受态GV3101中。

A.PH-START1基因扩增;B.阳性克隆PCR检测;C.pMD19- PH-START1双酶切验证;D.pSN1301-PH-START1的PCR检测;E.35S∶pSN1301-PH-START1双酶切验证;F.35S∶pSN1301-PH-START1转化GV3101后PCR检测。M.5 kb Marker;1-3.目的条带。
A. PH-START1 gene amplification; B. Positive clone detection by PCR; C.pMD19-PH-START1 double enzyme digestion test; D. pSN1301-PH-START1 PCR detection; E.35S∶pSN1301-PH-START1 double enzyme digestion test; F.PCR detection of pSN1301-PH-START1 transformation into GV3101. M. 5 kb Marker; 1-3. Target band.
图2 PH-START1过表达载体的构建
Fig.2 Construction of PH-START1 overexpressing vector
2.3 过表达PH-START1转基因拟南芥的遗传转化和筛选
用花絮侵染法转化拟南芥,将转化得到的种子经抗性筛选(图3-A),得到T3纯合株系,提取纯合株系DNA,可以扩增出目的条带(图3-B),说明过表达PH-START1转基因拟南芥创制成功。
2.4 过表达拟南芥植株中PH-START1的表达量分析
为了解PH-START1过表达拟南芥中PH-START1的表达情况,从得到的阳性植株中选取了2个转基因株系提取总RNA,反转录后对PH-START1基因进行SqRT-PCR分析(图4),结果发现2个转基因植株中pH-START1的表达量均高于野生型植株,表明确实为PH-START1过表达株系,将其分别命名为OE1、OE2。
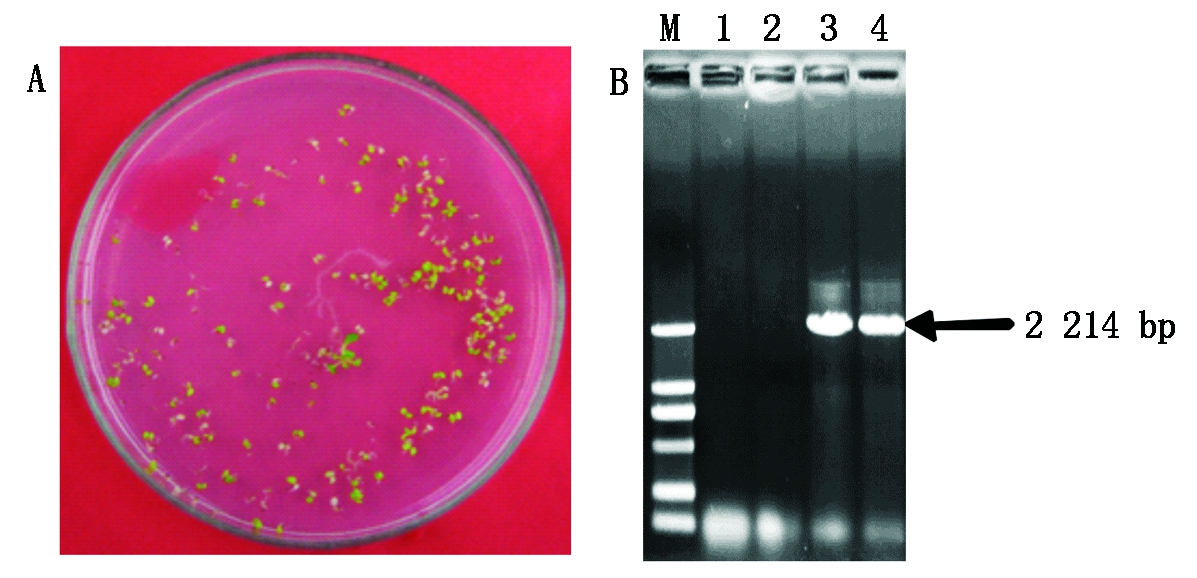
A.潮霉素平板上转基因阳性苗的筛选;B.PCR检测35S ∶pSN1301-PH-START1转基因阳性纯合株系。M.2 kb Marker;1-2.WT对照;3-4.目的条带。
A.Screening of transgenic positive seedlings on the hygromycin plate; B.PCR detection of the 35S∶pSN1301-PH-START1 transgenic positive homozygous lines. M.2 kb Marker;1-2.Control;3-4.Objective stripe.
图3 过表达PH-START1转基因阳性苗的筛选与纯合验证
Fig.3 Screening and homozygous validation of 35S∶pSN1301-PH-START1 overexpression transgenic positive seedlings
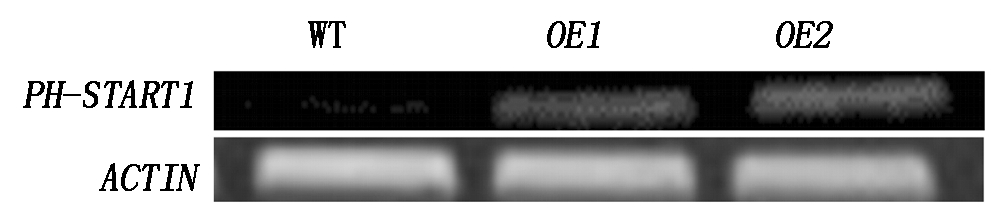
图4 SqRT-PCR分析PH-START1-OE转基因植株PH-START1表达量
Fig.4 The expression analysis of PH-START1 by SqRT-PCR in PH-START1-OE transgenic plants
2.5 PH-START1缺失突变体转基因拟南芥的创制
2.5.1 PH-START1 T-DNA插入突变体插入位点分析 由ABRC突变体库购得拟南芥PH-START1的2株不同T-DNA插入位点突变体,其编号分别为SALK_060171、SALK_120376。通过拟南芥数据库TAIR网站,得到其插入位点(图5),箭头所指的位置及方向就是载体插入的位置及方向。SALK_060171插入位置在起始密码子ATG后205 bp的位置,将之命名为ph-start1-1,SALK_120376插入位置在起始密码子ATG前534 bp的位置,将之命名为ph-start1-2,背景为 Col-0 野生型拟南芥。
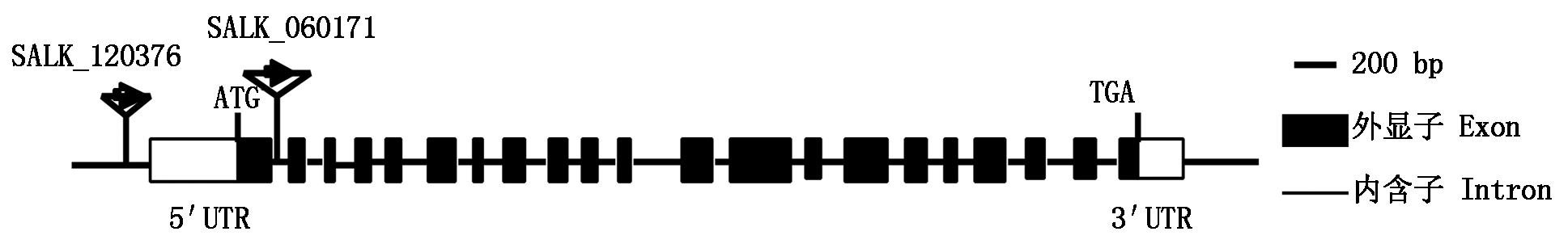
图5 ph-start1突变体中T-DNA插入位点示意图
Fig.5 The T-DNA insertion sites in ph-start1 mutant
提取T3不再出现分离比的突变体植株幼苗和野生型拟南芥的DNA,以T-DNA Primer Design设计插入位点上下游的引物LP、RP,以LBb1作为T-DNA特异引物,通过琼脂糖凝胶电泳验证T-DNA插入位点。结果表明(图6),ph-start1-1和ph-start1-2株系为T-DNA插入突变体纯合株系。
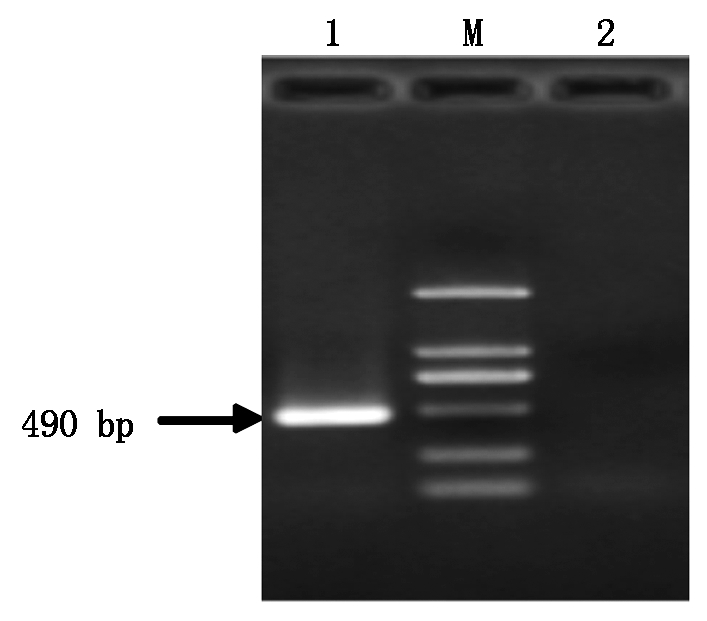
M.2 kb Marker;1.以LBb1和RP为引物;2.以LP和RP为引物。
M.2 kb Marker; 1.LBb1 and RP as primers; 2.LP and RP as primers.
图6 三引物法验证PH-START1 T-DNA插入位点
Fig.6 Validation of PH-START1 T-DNA insertion site by three primers method
2.5.2 T-DNA插入突变体中PH-START1的表达量分析 为了解PH-START1 T-DNA插入突变体中PH-START1表达情况,提取野生型拟南芥和ph-start1突变体幼苗RNA,用SqRT-PCR技术分析PH-START1基因的表达情况(图7),结果表明ph-start1-1和ph-start1-2无PH-START1表达,说明确实为PH-START1功能缺失突变体。
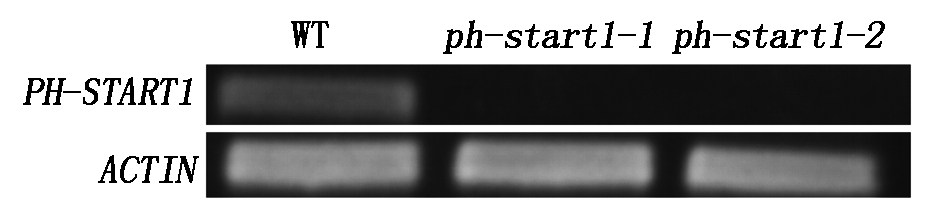
图7 SqRT-PCR技术分析PH-START1 T-DNA插入突变体中PH-START1的表达量
Fig.7 Expression analysis of PH-START1 by SqRT-PCR in PH-START1 T-DNA insertion mutants
2.6 PH-START1 转基因拟南芥表型分析
将收获的Col-0野生型拟南芥、PH-START1-OE、缺失突变体拟南芥种子播种在培养基中,放置于培养室相同环境下培养,观察各时期生长发育情况(图8)。发现播种12 d,Col-0野生型拟南芥为四叶期而OE转基因拟南芥已到达六叶期,OE植株明显较野生型大;播种25 d,OE生长发育已变得迟缓,植株明显小于野生型;播种40 d,OE转基因拟南芥各个器官的生长发育都明显小于野生型。ph-start1无论是在四叶期还是在营养生长时期的生长发育都略优于野生型。综上所述,PH-START1对拟南芥的生长发育起着重要的调控作用。

A.拟南芥四叶期植株表型对比(播种12 d);B.拟南芥四叶期植株表型对比(播种10 d);C-D.在同样正常的环境条件下,Col-0野生型和OE表型对比(播种25 d);E.Col-0野生型和ph-start1表型对比(播种30 d);F.Col-0野生型和OE表型对比(播种40 d)。
A.Phenotypic comparison of 4-leaf stage of Arabidopsis (12 d after sown); B.Phenotypic comparison of 4-leaf stage of Arabidopsis (10 d after sown); C-D.Phenotypic comparison of Col-0 wild type and OE (25 d after sown) under the same normal environmental conditions; E.Phenotypic comparison of Col-0 wild type and ph-start1 (30 d after sown); F.Phenotypic comparison of Col-0 wild type and OE (40 d after sown).
图8 PH-START1功能获得和缺失转基因拟南芥表型分析
Fig.8 Phenotypic analysis of PH-START1 transgenic Arabidopsis with functional acquisition and loss

A-B.OE拟南芥叶片的表型; C-D. ph-start1拟南芥叶片的表型;E.OE叶面积统计;F.ph-start1叶面积统计。
A-B.OE Arabidopsis leaves phenotype; C-D.ph-start1 Arabidopsis leaves phenotype; E.Comparison of leaf area of OE Arabidopsis;F.Comparison of leaf area of ph-start1 Arabidopsis.
图9 PH-START1转基因拟南芥叶的表型分析
Fig.9 Phenotype analysis of PH-START1 transgenic Arabidopsis leaves
观察相同培养条件下培养的OE和ph-start1的全部莲座叶,选择同一部位的莲座叶进行叶面积和叶柄长度的测定(图9)。结果发现,与野生型相比,OE的叶片极显著变小,叶面积为0.11,0.12 cm2,只占对照的4.89%,5.33%;ph-start1的叶片极显著变大,叶面积为2.94,3.05 cm2,占对照的142%,148%。表明PH-START1的过量表达严重影响了拟南芥叶片的发育,PH-START1表达量的下降可以促进叶片的发育。
3 讨论
拟南芥中PH-START1含有PH-START结构域,PH结构域首次被鉴定时认为是蛋白质结合域,现多参与细胞内信号传导、细胞骨架组织、膜转运和磷脂修饰[17-18]。START结构域首次被发现是在急性调节蛋白中,它是一个与脂类和甾醇类物质结合相关的结构域,负责将急性调节蛋白中的胆固醇转移到线粒体内膜[19-20]。在拟南芥中含START结构域的蛋白质家族共有35个成员,其中21个被融合在同源异型结构域中,此现象表明START结构域在植物的生长发育过程中起到了重要的作用[21-22]。随着拟南芥的生长发育,PH-START1在各个组织中均有不同程度的表达,根中主要在幼苗期表达量较高,莲座叶中主要在生长的第6片莲座叶表达量较高,花中主要在15花期表达量较高,角果中主要在授粉后5 d表达量较高,在花器官发育过程中,相对萼片、花瓣和雌蕊而言,雄蕊中PH-START1的表达量最高。
植物的生长周期分为2个阶段即营养生长阶段和生殖生长阶段。营养生长是生殖生长的基础,根系的发达才能从周围土壤中吸收大量的水分和营养物质,通过轴向和径向运输传递给茎和叶,使其茁壮生长,可以有效地抵制各种生物和非生物胁迫,并且为生殖生长做好准备。与野生型相比,过表达PH-START1拟南芥主根短、主茎细弱、莲座叶小,ph-start1的主根较长、主茎较粗、莲座叶大。PH-START1表达量的升高严重抑制了拟南芥的发育,说明PH-START1在拟南芥生长发育中起着负调控作用。至于PH-START1负调控拟南芥生长发育的分子机制,还需要对其进行转录组、蛋白质组及互作蛋白进行分析,建立植物生长发育调控网络,阐明植物生长发育调控机制。
[1] Ogawa E, Yamada Y, Sezaki N, Kosaka S A, Kamata N, Abe M, Komeda Y, Takahashi T.ATML1 and PDF2 play a redundant and essential role in Arabidopsis embryo development[J].Plant and Cell Physiology,2015,56(6):1183-1192.doi:10.1093/pcp/pcv045.
[2] Rombola-Caldentey B, Rueda-Romero P A, Carbonero P, Onate-Sanchez L. Arabidopsis DELLA and two HD-ZIP transcription factors regulate GA signaling in the epidermis through the L1 box cis-Element[J].Plant Cell,2014,26(7):2905-2919.doi:10.1105/tpc.114.127647.
[3] Zhu Y, Rong L, Luo Q, Wang B H, Zhou N A, Yang Y E, Zhang C, Feng H Y, Zheng L A, Shen W H, Ma J B, Dong A W. The histone chaperone NRP1 interacts with WEREWOLF to activate GLABRA2 in Arabidopsis root hair development[J].Plant Cell,2017,29(2):260-276.doi:10.1105/tpc.16.00719.
[4] Schrick K, Nguyen D, Karlowski W M, Mayer K F. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors[J].Genome Biology,2004,5(6):R41.doi:10.1186/gb-2004-5-6-r41.
[5] Tang D Z, Ade J, Frye C A, Innes R W. Regulation of plant defense responses in Arabidopsis by EDR2, a PH and START domain-containing protein[J].Plant Journal,2005,44(2):245-257. doi:10.1111/j.1365-313X.2005.02523.x.
[6] Shaw G.Identification of novel pleckstrin homology(PH)domains provides a hypothesis for PH domain function[J].Biochemical and Biophysical Research Communications, 1993, 195(2):1145-1151. doi:10.1006/bbrc.1993.2164.
[7] Sneha R, Pallavi M, Modeling S B. Dynamics and phosphoinositide binding of the pleckstrin homology domain of two novel PLCs:η1 and η2[J].Journal of Molecular Graphics & Modelling,2018(85):130-144. doi: 10.1016/j.jmgm.2018.07.012.
[8] Kang Y L, Kim B G, Kim S, Lee Y A.Inhibitory potential of flavonoids on Ptdlns(3,4,5)P3 binding with the phosphoinositide-dependent kinase 1 pleckstrin homology domain[J].Bioorganic & Medicinal Chemistry Letters,2017,27(3):420-426.doi:10.1016/j.bmc1.2016.12.051.
[9] Rebecchi M J, Scarlata S.Pleckstrin homology domains: A common fold with diverse functions[J].Annual Review of Biophysics and Biomolecular Structure,1998,27(4):503.doi:10.1146/annurev.biophys.27.1.503.
[10] Naughton F B, Kalli A C, Sansom M S. Modes of interaction of pleckstrin homology domains with membranes: toward a computational biochemistry of membrane recognition[J].Journal of Molecular Biology,2018,430(3):372-388.doi:10.1016/j.jmb.2017.12.011.
[11] Kalli A C, Campbell I D, Sansom M S. Interactions of the kindlin family pleckstrin homology domains with model membranes containing zwitterionic lipids and phosphatidyl inositol phosphates[J].Biophysical Journal,2014,106(2, 1):517A.doi:10.1016/j.bpj.2013.11.2889.
[12] Goraia S, Bagdia P R, Boraha R, Paulb D, Santrab M K, Khanc A T, Mannaa D. Insights into the inhibitory mechanism of triazole-based small molecules on phosphatidylinositol-4,5-bisphosphate binding pleckstrin homology domain[J].Biochemistry and Biophysics Reports, 2015, 2:75-86. doi: 10.1016/j.bbrep.2015.05.007.
[13] Panda P K, Behera B, Meher B R, Mukhopadhyay S, Sinha N A, Roy B, Das J, Paul S, Maiti T K, Agarwal R, Bhutia S K. Abrus agglutinin, a type Ⅱ ribosome inactivating protein inhibits Akt/PH domain to induce endoplasmic reticulum stress mediated autophagy-dependent cell death[J].Molecular Carcinogenesis,2017,56(2):389-401.doi:10.1002/mc.22502.
[14] Lemmon M A.Phosphoinositide recognition domains[J].Traffic,2003,4(4):201-213. doi:10.1034/j.1600-0854.2004.00071.x.
[15] Kumagai K, Elwell C A, Ando S, Engel J N, Hanada K.Both the N-and C-terminal regions of the Chlamydial inclusion protein D (IncD) are required for interaction with the pleckstrin homology domain of the ceramide transport protein CERT[J].Biochemical and Biophysical Research Communications,2018,505(4):1070-1076. doi:10.1016/j.bbrc.2018.09.168.
[16] 周苹. GH3.9基因过表达对拟南芥生长发育的影响研究[D]. 长沙: 湖南大学, 2013.doi: 10.7666/d.Y2523085.
Zhou P. Effect of over-expression og GH3.9 gene on the plant growth and development of Arabidopsis[D]. Changsha: Hunan University, 2013.
[17] Romanowski M J, Soccio R E, Breslow J L, Burley S K.Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain[J].Proceedings of the National Academy of Sciences of the United States of America,2002,99(10):6949-6954.doi:10.1073/pnas.052140699.
[18] Kang Y, Jang G, Ahn S, Lee Y A, Yoon Y. Regulation of AKT activity by inhibition of the pleckstrin homology Doniam-PtdIns(3,4,5)P-3 interaction using flavonoids[J].Journal of Microbiology and Biotechnology,2018,28(8):1401-1411.doi:10.4014/jmb.1804.04051.
[19] Yamada S, Yamaguchi T, Hosoda A, Iwawaki T, Kohno K. Regulation of human STARD4 gene expression under endoplasmic reticulum stress[J].Biochemical and Biophysical Research Communications,2006,343(4):1079-1085. doi:10.1016/j.bbrc.2006.03.051.
[20] Sluchanko N N, Tugaeva K V, Maksimov E G.Solution structure of human steroidogenic acute regulatory protein STARD1 studied by small-angle X-ray scattering[J].Biochemical and Biophysical Research Communications,2017,489(4):445-450.doi:10.1016/j.bbrc.2017.05.167.
[21] Kubo H, Peeters A J, Aarts M G, Pereira A, Koornneef M.ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis[J].The Plant Cell,1999,11(7):1217-1226. doi:10.2307/3870744.
[22] Schrick K, Bruno M, Khosla A, Cox P N, Marlatt S A, Roque R A, Nguyen H C, Snyder M P, Singh D, Yadav G.Shared functions of plant and mammalian StAR-related lipid transfer (START) domains in modulating transcription factor activity[J].BMC Biology,2014,12:70-78.doi:10.1186/s12915-014-0070-8.