燕麦(Avena sativa L.)是世界各地广泛栽培种植的一种重要的粮食兼饲草、饲料作物,其富含β-葡聚糖,被证明具有降血糖和血脂的功效[1],是美国FDA和英国JHCI认定的功能性谷物[2-3],随着燕麦及其加工产品的世界消费量的增长,传统燕麦育种技术已经不能有效满足燕麦生产的需要,并且成为制约燕麦育种进程的重要因素,而分子辅助育种是加速燕麦育种的重要手段。
目前,燕麦的育种多依赖于常规育种和表型鉴定研究,已成功育成了多种适应不同需求的燕麦品种并用于大田生产[4]。早期的表型特征量化研究证明燕麦具有丰富的遗传多样性[5],之后燕麦种质资源遗传连锁图[6]、皮裸性基因相关联SSR[7]和AFLP标记[8]也被相继报道。单核苷酸多态性(SNP)是目前最具优势的分子标记,被广泛应用于作物遗传图谱构建、QTL定位、全基因组关联分析(GWAS)、群体进化等[9-12]。近年来,基于下一代测序技术(Next generation sequencing,NGS)的基因分型技术(Genotyping by sequencing,GBS)是通过酶切处理去除基因组重复序列,显著降低了测序量和测序成本,且能够覆盖整个基因组,不受参考基因组限制,因而被广泛应用到开发高密度、高精度的农作物SNP分子标记研究中。目前,燕麦全基因组序列还没有发表,基因分型技术对农作物遗传多样性研究、种质资源鉴定和分子辅助育种标记开发具有重要的意义[13-16]。基因分型技术从单酶切技术RAD[17]、2b-RAD[18]、GBS[19]到现在的双酶切技术dd-GBS[20]、SLAF[21]均已被应用到玉米、小麦等[22-30]主要农作物分子标记开发研究。Winkler 等[31]采用GBS技术,证明了燕麦颖片颜色和穗型等表型特征和SNP位点相关。在另一项研究中,通过GBS技术构建的高密度SNP则能精准地区分起源于世界不同地区燕麦种质资源[32],证实GBS-SNP技术是种质资源鉴定的有力工具。
本研究基于NGS的dd-GBS技术,以经过抗旱性鉴定的燕麦种质为试验材料,选用适合燕麦基因组酶切的酶,运用适合燕麦简化基因组参考序列的分析软件,在构建燕麦简化基因组参考序列的基础上,研究燕麦相关SNP标记,旨在丰富当前燕麦的基因组数据库,提供分子标记(SNP)基础数据,为加速燕麦育种进程提供理论支撑。
1 材料和方法
1.1 试验材料
供试材料由中国农业科学院品种资源研究所麦类室于1981-1983年进行抗旱鉴定的燕麦种质42份,包括11份高抗(High tolerance)、13份中抗(Moderate tolerance)、18份低抗(Low tolerance)和山西省农业科学院农作物品种资源研究所燕麦种质资源圑提供[33]的抗旱性未鉴定(Uncharacterized)燕麦育成品种(品系)12个(表1),2016年种植于山西省农业科学院智能温室。
1.2 试验方法
GBS文库构建与数据质控:燕麦种子发芽14 d,取鲜嫩叶片提取基因组DNA(Mag-MK Plant Genomic DNA Extraction Kit试剂盒,上海生工),1.0%琼脂糖凝胶电泳和紫外分光光度计分别检测DNA质量。用限制性内切酶PstⅠ(CTGCAG)和 MspⅠ(CCGG)对提取的DNA 进行酶切后,回收大小在220~450 bp的酶切片段,之后按照dd-GBS方法进行建库[20]。采用llumina Hiseq测序平台进行双末端(Paired-end,PE)测序,获得的下机数据以双端FASTQ格式保存。
分别对每个样品的下机数据运用 FastQC软件进行质量控制,主要包括碱基质量分布(Per base sequence quality)、质量值(Per sequence quality scores)、GC分布(Per sequence GC content),对反映下机数据质量的多项指标进行统计。进一步过滤数据包含一些带接头,双末端reads1 5′端6 bp非酶切位点序列CTGCAG或reads2的5′端4 bp非酶切位点序列CCGG。采用Adapter Removal去除3′端的接头污染[34]。采用滑动窗口进行质量过滤,窗口大小设置为5 bp,步长设置为1 bp,每一次往前移动1个碱基,取5个碱基计算窗口的平均Q值,若最后一个碱基的Q值≤2,则仅保留该位置之前的碱基;若窗口的平均Q值≤20,则仅保留该窗口倒数第2个碱基及之前的碱基。滤除小于50 bp 的reads。
表1 不同抗旱性的燕麦试验材料
Tab.1 Sequenced oats germplasm of different drought tolerance
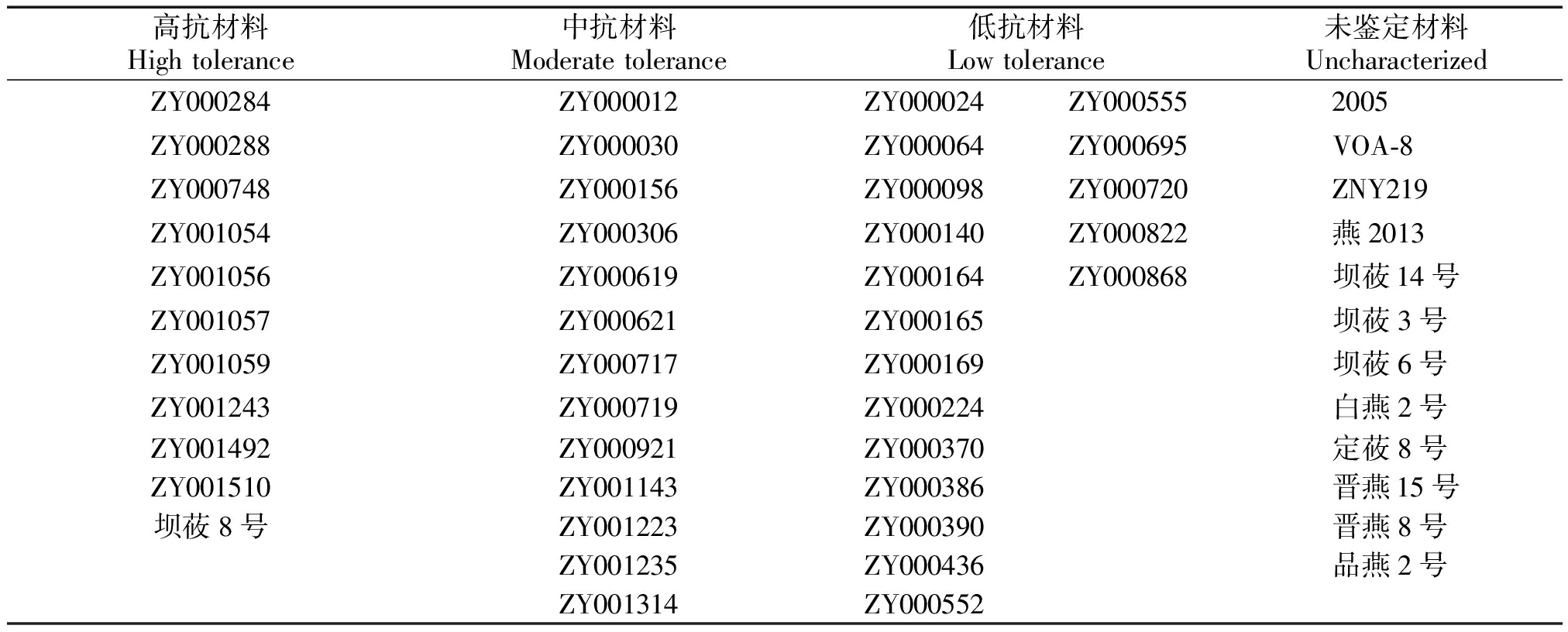
高抗材料High tolerance中抗材料Moderate tolerance低抗材料Low tolerance未鉴定材料UncharacterizedZY000284ZY000012ZY000024ZY0005552005ZY000288ZY000030ZY000064ZY000695VOA-8ZY000748ZY000156ZY000098ZY000720ZNY219ZY001054ZY000306ZY000140ZY000822燕2013ZY001056ZY000619ZY000164ZY000868坝莜14号ZY001057ZY000621ZY000165坝莜3号ZY001059ZY000717ZY000169坝莜6号ZY001243ZY000719ZY000224白燕2号ZY001492ZY000921ZY000370定莜8号ZY001510ZY001143ZY000386晋燕15号坝莜8号ZY001223ZY000390晋燕8号ZY001235ZY000436品燕2号ZY001314ZY000552
1.3 数据分析
1.3.1 燕麦简化基因组参考序列组装与群体SNP检测 应用Stacks软件包[35]中的ustacks对每个样品的reads进行聚类,同一个stack代表1个酶切位点(loci),聚类参数-m设置为4,对每一个样品的loci及loci的测序深度进行统计。下一步用cstacks将所有样品的loci合并,不同样品loci之间最多允许2个错配,获得每个loci的catalog consensus序列。
采用sstacks将每个样品的loci序列与catalog consensus序列比对后populations过滤获得群体SNP。主要参数包括:1个位点最少要在1个群体中出现;1个群体中检测到同一位点的个体最低百分数50%(当群体中该位点的缺失率超过 50%,则去除该位点);1个位点的最小等位基因频率0.05。
1.3.2 燕麦SNP分析 采用 GCTA软件利用群体SNP数据进行主成分聚类分析;对高、低抗材料的SNP数据进行Fisher Test统计分析获取相关SNP标记;采用结合校正标准错误发现率(False Discovery Rate,FDR)小于0.05和0.001确定差异显著性标记和极显著性标记。
1.3.3 燕麦SNP功能预测 进一步对差异极显著(FDR<0.001)的SNP所在源序列和小麦(Triticum aestivum L.)基因组序列进行Blast比对,筛查SNP相关基因。
2 结果与分析
2.1 燕麦GBS测序结果
本研究共测序54个样本,注释reads数在4 111 218~21 782 382;质控后保留的高质量数据(reads数4 111 218~19 019 296)用于后续的数据分析(图1)。
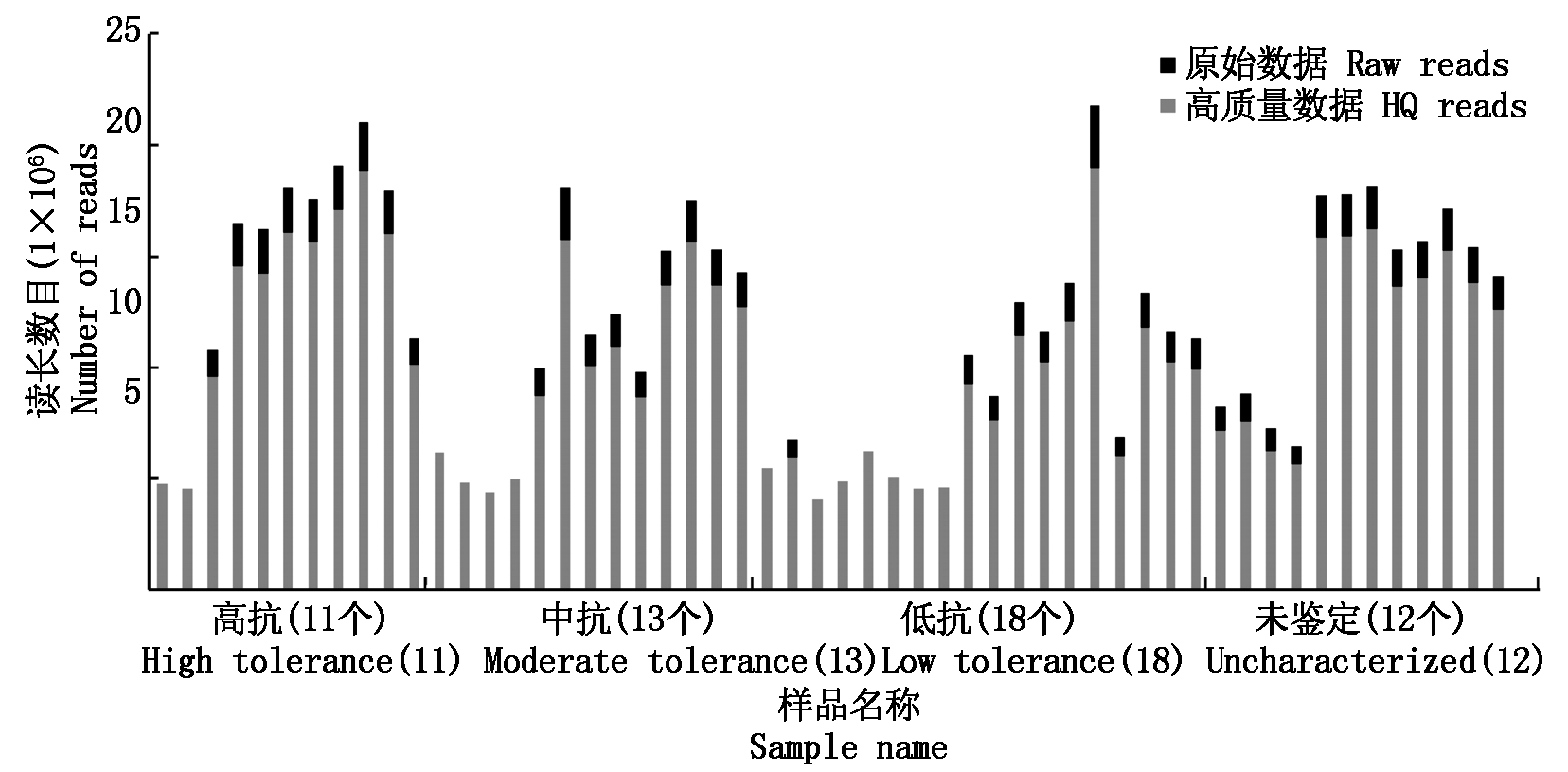
图1 燕麦GBS-SNP注释序列
Fig.1 Oat GBS-SNP annotation sequence
2.2 燕麦参考序列与群体SNP位点
采用Stacks数据包对燕麦reads进行聚类,结果显示:每个loci平均测序深度为20.61×,合并所有样本同一个loci,共获得753 325个参考基因组序列(catalog consensus)。每个样品的loci与其对应的catalog比对,去除群体位点缺失(./.)率超过50%和最小等位基因频率小于0.05的SNP位点,共获得74 657个群体SNP位点。由图2可知,所有测序样本中纯合状态的SNP位点(0/0或1/1)占少数,大部分呈现不同程度的杂合(0/1)或缺失状态(./.)。

0/0 和1/1.纯合体;0/1.杂合体;./..缺失。
0/0 和1/1. Homozygote; 0/1.Heterozygote;./.. Deletion.
图2 SNP位点杂合性
Fig.2 SNP heterogeneity
2.3 燕麦SNP研究
根据群体SNP数据,对54份燕麦种质的PCA结果显示,SNP基因型明显聚类为2簇,全部11份高抗、4份抵抗、8份中抗种质和6个未鉴定品种聚为一簇;而另外一簇则包含14份低抗和5份中抗种质;其余6个未鉴定的燕麦品种则分布在2簇之外(图3)。
基于PCA基因型聚类结果,进一步对高抗(n=11)和低抗(n=18)种质2组进行SNP基因型差异显著分析,共获得2 937个燕麦抗旱性相关SNP(Fisher Test,FDR<0.05)。其中,差异极显著(FDR<0.001)的55个SNP位点坐落于41个燕麦简化基因组参考序列(同一参考序列包含2或3个SNP位点)。这55个相关SNP可分为3类,23个SNP 位点在高抗种质中缺失; 28个SNP位点在低抗种质中缺失;其余4个SNP位点在2种种质中都存在(表2)。
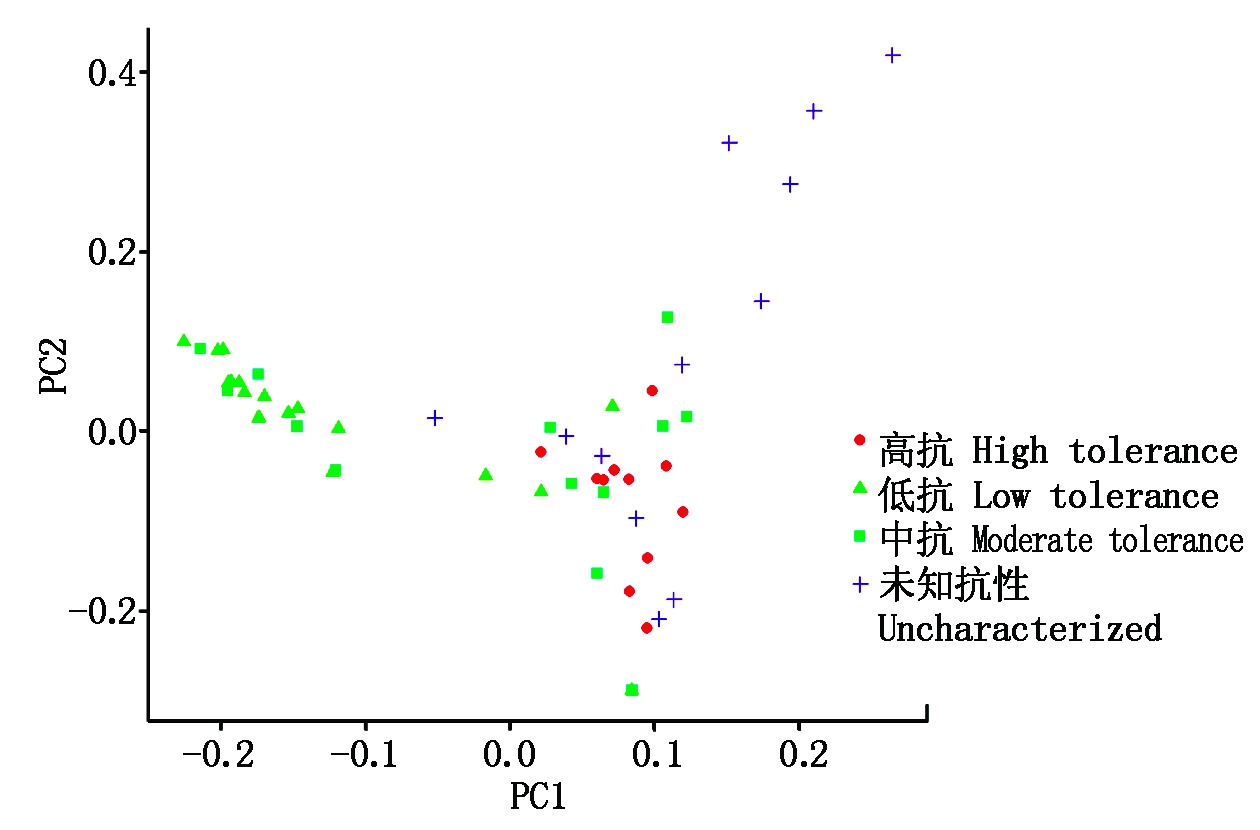
图3 燕麦SNP基因型主成分分析
Fig.3 Principal component analysis of oat SNP genotypes
表2 高抗与低抗燕麦的差异显著SNP位点
Tab.2 Oat significant SNPs of high tolerance and low tolerance

SNP序列号Catalog位点Position类型Type高抗材料( n=11)High tolerance (n=11)低抗材料(n=18)Low tolerance (n=18)cat 5909843C>GCC(11)CC(2)、 GG(15)、 ▲(1)cat 27798992G>AGG(11)GG(2)、 AA(14)、 ▲(2)cat 325976126C>ACC(11) CC(2)、 AA(14)、 ▲(2)cat 11935694G>AAA(9)、 GA(1)、 ▲(1)GG(8)、 GA(7)、 ▲(3)cat 93171135A>G▲(11)AG(18)cat 18630383G>T▲(11)GT(18)cat 31736230G>T▲(11)GG(18)cat 31736260C>G▲(11)CC(18)
表2(续)

SNP序列号Catalog位点Position类型Type高抗材料( n=11)High tolerance (n=11)低抗材料(n=18)Low tolerance (n=18)cat 29746449G>A▲(11)GG(4)、 GA(12)、 AA(1)、 ▲(1)cat 297464112G>A▲(11)GG(4)、 GA(12)、 AA(1)、 ▲(1)cat 297464115G>A▲(11)GG(4)、 GA(11)、 AA(1)、 ▲(2)cat 5058688C>A▲(11)CC(1)、 CA(14)、 AA(2)、 ▲(1)cat 29253583T>C▲(11)TT(2)、 TC(12)、 CC(1)、 ▲(3)cat 29253598C>T▲(11)CC(2)、 CT(12)、 TT(1)、 ▲(3)cat 292535119A>T▲(11)AA(2)、 AT(12)、 TT(1)、 ▲(3)cat 16254029C>T▲(11)CC(2)、 CT(11)、 ▲(5)cat 239321108T>C▲(11)TT(15)、 TC(2)、 ▲(1)cat 69705135A>C▲(11)AA(2)、 CC(14)、 ▲(2)cat 31126332G>A▲(11)GG(2)、 GA(10)、 ▲(6)cat 31126349C>T▲(11)CC(2)、 CT(10)、 ▲(6)cat 31126374T>C▲(11)TT(2)、 TC(10)、 ▲(6)cat 15216716T>G▲(11)TT(13)、 TG(1)、 ▲(4)cat 15216798G>A▲(11)GG(13)、 GA(1)、 ▲(4)cat 152167100A>G▲(11)AA(13)、 AG(1)、 ▲(4)cat 2690133A>T▲(11)AA(5)、 AT(8)、 ▲(5)cat 29274854C>T▲(11)CC(16)、 ▲(2)cat 2128933C>A▲(11)CC(8)、 CA(7)、 ▲(3)cat 19280561C>TCC(2)、 CT(7)、 TT(1)、 ▲(1)▲(18)cat 2023018A>GAA(1)、 AG(8)、 GG(1)、 ▲(1)▲(18)cat 20230196T>ATT(1)、 TA(8)、 AA(1)、 ▲(1)▲(18)cat 44134103T>CTT(1)、 TC(7)、 CC(1)、 ▲(2)▲(18)cat 203256111C>TCC(3)、 CT(3)、 TT(4)、 ▲(1)▲(18)cat 312272121A>GAA(1)、 AG(7)、 GG(1)、 ▲(2)▲(18)cat 10076739G>CGG(1)、 GC(8)、 ▲(2)▲(18)cat 127357132C>TCC(5)、 CT(5)、 ▲(1)▲(18)cat 23652378C>TCC(3)、 CT(7)、 TT(1)▲(18)cat 1319747G>CGG(8)、 GC(1)、 ▲(2)▲(18)cat 16836455A>GAA(1)、 AG(7)、 ▲(3)▲(18)cat 168364104T>CTT(1)、 TC(7)、 ▲(3)▲(18)cat 32726376T>CTT(7)、 TC(2)、 ▲(2)▲(18)cat 7199517G>TGG(1)、 GT(6)、 ▲(4)▲(18)cat 7199582A>CAA(1)、 AC(6)、 ▲(4)▲(18)cat 1726767T>CTT(8)、 TC(1)、 ▲(2)▲(18)cat 23615018G>AGG(3)、 GA(6)、 ▲(2)▲(18)cat 23615082G>AGG(3)、 GA(6)、 ▲(2)▲(18)cat 18288741G>TGG(10)、 GT(1)▲(18)cat 182887116A>CAA(10)、 AC(1)▲(18)cat 1757047A>CAA(9)、 AC(2)▲(18)cat 205608134C>TCC(9)、 CT(2)▲(18)cat 2471956C>TCC(8)、 CT(3)▲(18)cat 10599799C>GCC(6)、 CG(5)▲(18)cat 273019129G>CGG(10)、 GC(1)▲(18)cat 2995457G>CGG(9)、 GC(2)▲(18)cat 6077781C>ACA(8)、 ▲(3)▲(18)cat 24851130G>AGA(9)、 ▲(2)▲(18)
注:▲.SNP位点缺失。
Note: ▲. Represents site deletion of SNP.
2.4 燕麦SNP的Blast功能预测
进一步对差异极显著的55个相关SNP的源序列与小麦基因组序列进行了Blast比对,结果显示,10个源序列(包括14个SNP位点)和多个基因转录序列联配(表3)。其中,参与线粒体活性氧信号转导、植物激素信号转导的调控蛋白、自噬基因表达与植物抗逆、抗旱水平有关。
表3 燕麦SNP源序列联配基因(比对小麦基因组)
Tab.3 Oat SNPs alignment genes (against wheat genome)
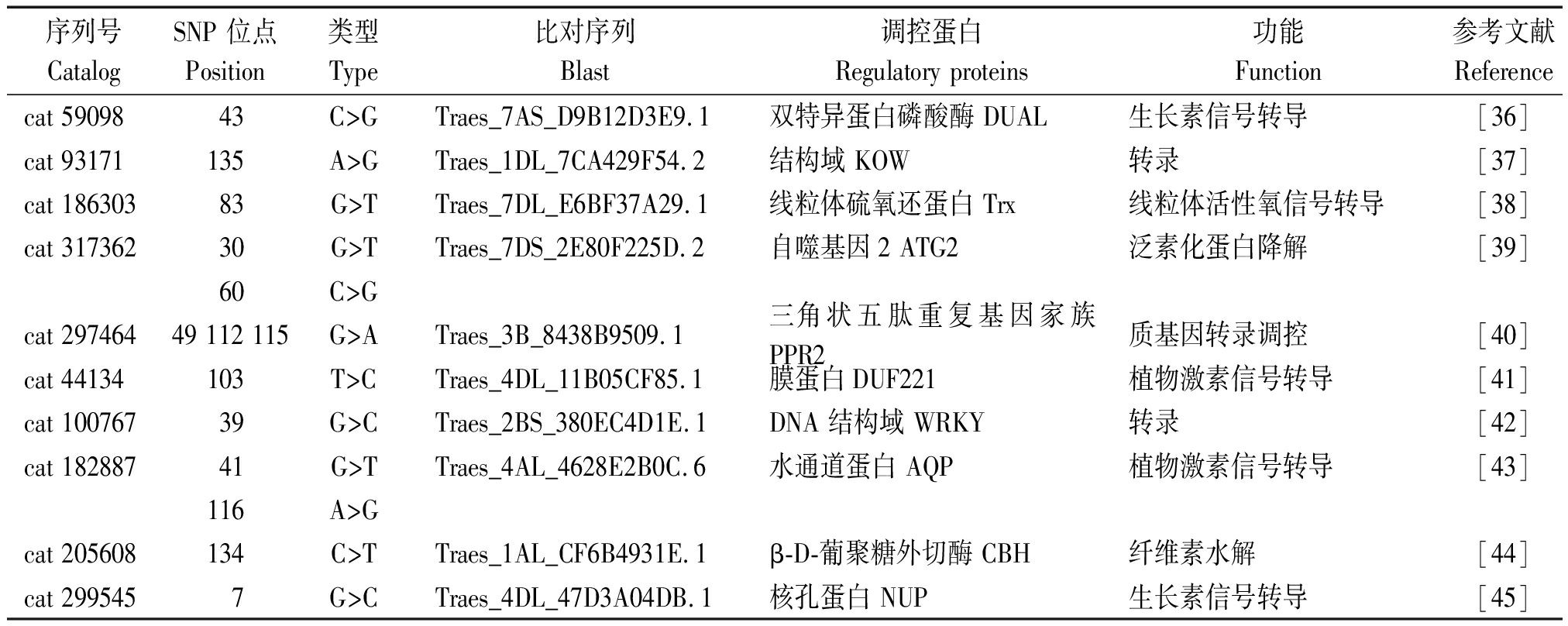
序列号CatalogSNP 位点Position类型Type比对序列Blast调控蛋白Regulatory proteins功能Function参考文献Referencecat 5909843 C>GTraes_7AS_D9B12D3E9.1双特异蛋白磷酸酶 DUAL生长素信号转导[36]cat 93171135 A>GTraes_1DL_7CA429F54.2结构域 KOW转录[37]cat 18630383 G>TTraes_7DL_E6BF37A29.1线粒体硫氧还蛋白 Trx线粒体活性氧信号转导[38]cat 31736230 G>TTraes_7DS_2E80F225D.2自噬基因2 ATG2泛素化蛋白降解[39]60 C>Gcat 29746449 112 115G>ATraes_3B_8438B9509.1 三角状五肽重复基因家族 PPR2质基因转录调控[40]cat 44134103 T>CTraes_4DL_11B05CF85.1膜蛋白DUF221植物激素信号转导[41]cat 10076739 G>CTraes_2BS_380EC4D1E.1 DNA 结构域 WRKY转录[42]cat 18288741 G>TTraes_4AL_4628E2B0C.6 水通道蛋白 AQP植物激素信号转导[43]116 A>Gcat 205608134 C>TTraes_1AL_CF6B4931E.1β-D-葡聚糖外切酶 CBH纤维素水解[44]cat 2995457 G>CTraes_4DL_47D3A04DB.1核孔蛋白 NUP生长素信号转导[45]
3 结论与讨论
3.1 GBS技术开发高密度SNP分子标记
高通量测序技术的面世是现代分子生物学技术的革命性飞跃,NGS技术促使生命科学研究进入基因组学时代,NGS在农作物上的重要应用之一就是GBS技术。GBS技术已经成为加速农作物种质资源鉴定筛选以及分子辅助育种的重要技术之一[16,19,46],燕麦基因组相对复杂(2n=6x=42),具有丰富的遗传多样性。本研究采用新一代ddGBS技术,成功组装了基于54个样本的燕麦简化基因组参考序列。质控显示,获得的高质量reads通量和SNP杂合状态与抗旱性没有明显关联。GBS通过酶切过滤重复序列,降低基因组复杂程度,虽不代表全基因组序列结构,但非常适合于农作物分子辅助育种SNP标记开发。本研究之所以选择经过多年抗旱性鉴定、遗传稳定、抗旱梯度差异明显(高抗、中抗、低抗)的燕麦种质资源进行测序,目的是通过抗旱性表型与SNP基因型关联分析,开发可能与抗旱性有关联的SNP。因本研究样本数量相对偏小(n=54),在构建简化基因组参考序列中没有采用报道的针对大样本TASSEL分析流程[33],而应用更适合于小群体的Stacks分析流程[35]。虽技术路线不同,但每项研究贡献的简化基因组参考序列无疑丰富了目前的燕麦基因组数据库。
3.2 抗旱性和SNP基因型关联分析
本研究的主要目的是挖掘可能与燕麦抗旱性有关联的SNP,为燕麦种质资源鉴定和分子辅助育种服务。这些试验材料根据多年的抗旱性鉴定划分为高抗、中抗和低抗3组,但根据SNP基因型聚类分析则大致分为2簇,高抗材料SNP基因型和大多数低抗材料基因型各聚类为一簇,明确显示抗旱性和SNP基因型是关联的,也就是说SNP基因型很大程度上能反映抗旱性。中抗材料SNP基因型没有聚类成簇,而是呈分散状态,也从侧面说明表型是基因型与环境共同作用的结果,同时作物抗旱性又是一个多指标的综合性状表现,但基因型的选择是改良作物性状的有效途径[47-48]。燕麦抗旱表型是抗旱基因决定的性状在环境作用下的具体表现,其抗旱性鉴定结果可能在不同的环境条件下会有所偏差。
因高、低抗材料的抗旱性和SNP基因型相关联,本研究针对这2组材料,开发可能与燕麦抗旱性相关的SNP,大部分SNP属于杂合或缺失状态,只有极少数呈现纯合状态,可能是因为燕麦基因组高度杂合(六倍体)以及测序技术的原因。下一步仍需要应用精度更高的数字化 PCR 技术在更大群体验证本研究发现的SNP。
利用GBS技术开发可能与燕麦抗旱性相关SNP标记在其他作物育种研究中已经报道[49-53]。对于燕麦来讲,本研究采用GBS技术,第1个报道可能与燕麦抗旱性有关联的SNP标记,以期为燕麦种质资源精准筛查和分子辅助育种提供数据基础。
3.3 相关SNP生物学功能
基因组技术已被广泛应用到抗旱基因挖掘[54-56]。GRLs[57]、OsMYB55 [58]和GIF[59] 被相继报道与不同作物抗旱机制有关。本研究经Blast比对获得的相关SNP源序列和不同生物通路基因转录子联配。其中cat 186303的第83个碱基在高抗材料中位点缺失,而在低抗材料中则呈现GT杂合状态。cat 186303源序列Blast 比对小麦基因组数据库显示,其序列与小麦硫氧还蛋白Trx序列相似。Trx蛋白参与细胞生长凋亡和基因转录[60],多项研究证明,Trx基因和作物抗旱、抗逆功能有关[16, 61-65]。后续功能验证包括研究 cat 186303对燕麦Trx基因表达调控,是解释功能SNP的重要步骤。此外,其他抗旱性相关SNP源序列联配的基因包括植物激素信号转导的DUF221、双特异蛋白磷酸酶和核孔蛋白,与植物抗逆有关。
3.4 本研究的局限性
燕麦是六倍体作物,本研究所开发的燕麦抗旱相关联SNP不能定位,但证明不同抗性材料的燕麦在SNP存在表达差异,是作为燕麦材料分子筛选的潜在标记。因为抗旱性状是一个综合指数,单一参数不能全面代表抗旱能力。本试验得出的特异性SNP 需要今后在大量的群体材料中进行验证。此外,燕麦抗旱指标有待量化也是今后挖掘抗旱SNP 的重要基础。本研究结果支持表型-基因型互作关系的同时也指出,抗旱性是基因型和环境因子综合体现,初期开发的可能与燕麦抗旱性有关联的SNP必须在更大种质群体中验证以加强其SNP的广谱性和实用性。
常规育种主要依赖于表型数据,而分子辅助育种则侧重于基因型数据,构建燕麦抗旱性表型-基因型关联则是精准育种的必经之路。应用GBS技术,本研究成功组装了燕麦简化基因组参考序列并初步研究了可能与燕麦抗旱性有关联的SNP标记,研究结果不仅丰富了燕麦基因组数据库,同时为燕麦种质资源早期筛选和燕麦基因数据库提供数据基础。
致谢:感谢荷兰莱顿大学统计系Goeman J J教授的统计学方法指导;感谢美国加州戴维斯分校植物系罗明诚教授对本试验给予的建议。
[1] Wang Q, Ellis P R.Oat beta-glucan: physico-chemical characteristics in relation to its blood-glucose and cholesterol-lowering properties[J].British Journal of Nutrition,2014,112(2):S4-S13.doi:10.1017/S0007114514002256.
[2] 任顺成, 马瑞萍. 燕麦的功效因子及其保健功能[J]. 粮食科技与经济, 2013, 38(3): 58-60. doi:10.16465/j.gste.2013.03.015.
Ren S C, Ma R P. Efficacy factor and health care function of oat[J]. Grain Science and Technology and Economy, 2013,38(3):58-60. doi:10.16465/j.gste.2013.03.015.
[3] Herranen M, Kariluoto S, Edelmann M, Piironenb V, Ahvenniemib K, Iivonenb V, Salovaarab H, Korhola M, et al.Isolation and characterization of folate-producing bacteria from oat bran and rye flakes[J].International Journal of Food Microbiology,2010,142(3):277-285. doi.org/10.1016/j.ijfoodmicro.2010.07.002.
[4] 崔林, 杨才, 任长忠, 付晓峰, 张宗文, 乔治军, 范银燕, 刘根科, 韩美善, 刘龙龙. 燕麦育种技术基础研究及新品种选育[J]. 中国科技成果, 2014(10): 21-23. doi:10.3772/j.issn.1009-5659.2014.10.009.
Cui L, Yang C, Ren C Z, Fu X F, Zhang Z W, Qiao Z J, Fan Y Y, Liu G K, Han M S, Liu L L. Basic research on oat breeding technology and breeding of new varieties[J]. China Science and Technology Achievements, 2014(10):21-23. doi:10.3772/j.issn.1009-5659.2014.10.009.
[5] Diederichsen A.Assessments of genetic diversity within a world collection of cultivated hexaploid oat (Avena sativa L.) based on qualitative morphological characters[J].Genetic Resources and Crop Evolution,2008,55(3):419-440.doi:10.1007/s10722-007-9249-y.
[6] 徐微, 张宗文, 吴斌, 崔林. 裸燕麦种质资源AFLP标记遗传多样性分析[J]. 作物学报, 2009, 35(12): 2205-2212. doi:10.3724/SP.J.1006.2009.02205.
Xu W, Zhang Z W, Wu B, Cui L. Genetic diversity in naked oat (Avena nuda) germplasm revealed by AFLP markers[J]. Acta Agronomica Sinica, 2009, 35(12):2205-2212. doi:10.3724/SP.J.1006.2009.02205.
[7] 王玉亭. 燕麦籽粒皮裸性基因遗传与分子作图[D]. 北京: 中国农业科学院, 2011.
Wang Y T. Genetic and molecular mapping of naked genes in oat seed skin[D]. Beijing: The Chinese Academy of Agricultural Sciences, 2011.
[8] 吴斌, 张茜, 宋高原, 陈新, 张宗文. 裸燕麦SSR标记连锁群图谱的构建及β-葡聚糖含量QTL的定位[J]. 中国农业科学, 2014, 47(6): 1208-1215. doi: 10.3864/j.issn.0578-1752.2014.06.017.
Wu B, Zhang Q, Song G Y, Chen X, Zhang Z W. Construction of SSR genetic linkage map and analysis of QTLs related to β-glucan content of naked oat(Avena nuda L.)[J]. Scientia Agricultura Sinica, 2014,47(6):1208-1215. doi: 10.3864/j.issn.0578-1752.2014.06.017.
[9] Moumouni K H, Kountche B A, Jean M, Hash C, Haussmann B, Belzile F. Construction of a genetic map for pearl millet, Pennisetum glaucum (L.) R. Br., using a genotyping-by-sequencing (GBS) approach[J].Molecular Breeding,2015,35(1):5.doi:10.1007/s11032-015-0212-x.
[10] Lin M, Cai S, Wang S, Liu S, Zhang G, Bai G.Genotyping-by-sequencing (GBS) identified SNP tightly linked to QTL for pre-harvest sprouting resistance[J]. Theoretical and Applied Genetics,2015,128(7):1385-1395. doi:10.1007/s00122-015-2513-1.
[11] Tumino G, Voorrips R E, Rizza F, Badeck F W, Morcia C, Ghizzoni R, Germeier C U, Paulo M J, Terzi V, Smulders M J. Population structure and genome-wide association analysis for frost tolerance in oat using continuous SNP array signal intensity ratios[J].Theoretical and Applied Genetics,2016,129(9):1711-1724.doi:10.1007/s00122-016-2734-y.
[12] Nawaz M Y, Jimenez-Krassel F, Steibel J P, Lu Y, Baktula A, Vukasinovic N, Neuder L, Ireland J, Ireland J J, Tempelman R. Genomic heritability and genome-wide association analysis of anti-M llerian hormone in Holstein dairy heifers[J].Journal of Dairy Science,2018,101(9): 1425-1430. doi:org/10.3168/jds.2018-14798.
[13] Torkamaneh D, Laroche J, Belzile F.Genome-wide SNP calling from genotyping by sequencing(GBS)data:a comparison of seven pipelines and two sequencing technologies[J].PLoS One,2016,11(8):e0161333. doi: 10.1371/journal.pone.0161333.
[14] Peterson G W, Dong Y, Horbach C, Yong B F.Genotyping-by-sequencing for plant genetic diversity analysis:a Lab guide for SNP genotyping[J].Diversity,2014,6(4):665-680. doi:org/10.3390/d6040665.
[15] Rasheed A, Hao Y F, Xia X C, Khan A A, Varshney R K, He Z H.Crop breeding chips and genotyping platforms: progress, challenges, and perspectives[J].Molecular Plant,2017,10(8):1047-1064.doi:10.1016/j.molp.2017.06.008.
[16] Scheben A, Batley J, Edwards D. Genotyping-by-sequencing approaches to characterize crop genomes: choosing the right tool for the right application[J]. Plant biotechnology Journal, 2017, 15(2): 149-161. doi:org/10.1111/pbi.12645.
[17] Miller M R, Dunham J P, Amores A, Cresko W A, Johnson E A. Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA(RAD) markers[J]. Genome Research, 2007,17(2), 240-248. doi:10.1101/gr.568120.
[18] Wang S, Meyer E, Mckay J K, Matz M V.2b-RAD: a simple and flexible method for genome-wide genotyping[J].Nature Methods,2012,9(8):808.doi:10.1038/NMETH.2023.
[19] Elshire R J, Glaubitz J C, Sun Q, Poland J A, Kawamoto K, Buckler E S, Mitchell S E.A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species[J].PLoS One,2011,6(5):e19379.doi:10.1371/journal.pone.0019379.
[20] Peterson B K, Weber J N, Kay E H, Fisher H S, Hoekstra H E.Double digest RADseq: an inexpensive method for De Novo SNP discovery and genotyping in model and Non-Model species[J].PLoS One,2012,7(5):0037135.doi:10.1371/journal.pone.0037135.
[21] Sun X W, Liu D Y, Zhang X F, Li W A, Hong W G, Jiang C B, Guan N, Ma C X, Zeng H P, Xu C H, Song J, Huang L, Wang C M, Shi J J, Wang R, Zheng X A, Wang X W, Zheng H K.SLAF-seq: an efficient method of large-scale De Novo SNP discovery and genotyping using high-throughput sequencing[J].PLoS One,2013,8(3):0058700.doi:10.1371/journal.pone.e58700.
[22] Alipour H, Bihamta M R, Mohammadi V A, Bai G H, Zhang G R. Genotyping-by-Sequencing (GBS) revealed molecular genetic diversity of iranian wheat landraces and cultivars[J].Frontiers in Plant Science,2017,8:01293.doi:10.3389/fpls.2017.01293.
[23] Annicchiarico P, Nazzicari N, Wei Y L, Pecetti L, Brummer E C. Genotyping-by-sequencing and its exploitation for forage and cool-season grain legume breeding[J].Frontiers in Plant Science,2017,8:679.doi:10.3389/fpls.2017.00679.
[24] Chang C W, Wang Y H, Tung C W.Genome-wide single nucleotide polymorphism discovery and the construction of a high-density genetic map for melon (Cucumis melo L.) using genotyping-by-sequencing[J].Frontiers in Plant Science,2017,8:125.doi:10.3389/fpls.2017.00125.
[25] Ertiro B T, Semagn K, Das B, Olsen M, Labuschagne M, Worku M, Wegary D A, Ogugo V, Keno T, Abebe B A, Menkir A.Genetic variation and population structure of maize inbred lines adapted to the midaltitude sub-humid maize agro-ecology of Ethiopia using single nucleotide polymorphic (SNP) markers[J].BMC Genomics,2017,18(1):777.doi:10.1186/s12864-017-4173-9.
[26] Gouesnard B, Negro S, Laffray A, Glaubitz J, Melchinger A, Revilla P, Moreno-Gonzalez J, Madur D, Combes V, Tollon-Cordet C, Laborde J, Kermarrec D, Bauland C, Moreau L, Charcosset A, Nicolas S. Genotyping-by-sequencing highlights original diversity patterns within a European collection of 1191 maize flint lines, as compared to the maize USDA genebank[J].Theoretical and Applied Genetics,2017,130(10):2165-2189. doi:10.1007/s00122-017-2949-6.
[27] Jo J, Purushotham P M, Han K, Lee H R, Nah G, Kang B C.Development of a genetic map for onion (Allium cepa L.) using reference-free genotyping-by-sequencing and SNP assays[J].Frontiers in Plant Science,2017,8:1606.doi:10.3389/fpls.2017.01606.
[28] Kumar S, Kirk C, Deng C, Wiedow C. Genotyping-by-sequencing of pear (Pyrus spp.) accessions unravels novel patterns of genetic diversity and selection footprints[J]. Horticulture research, 2017, 4: 17015. doi: 10.1038/hortres.2017.15.
[29] Malmberg M M, Pembleton L W, Baillie R C, Sudheesh S, Kaur S A, Verma P, Spangenberg G C, Forster J W, Cogan N O. Genotyping-by-sequencing through transcriptomics: implementation in a range of crop species with varying reproductive habits and ploidy levels[J].Plant Biotechnology Journal,2018,16(4):877-889.doi:10.1111/pbi.12835.
[30] Otto L G, Mondal P, Brassac J, Preiss S, Degenhardt J, He S, Reif J C, Sharbel T F.Use of genotyping-by-sequencing to determine the genetic structure in the medicinal plant chamomile,and to identify flowering time and alpha-bisabolol associated SNP-loci by genome-wide association mapping[J].BMC Genomics,2017,18(1):599. doi:org/10.1186/s12864-017-3991-0.
[31] Winkler L R, Bonman J M, Chao S A, Bockelman H, Klos K E.Population structure and genotype phenotype associations in a collection of oat landraces and historic cultivars[J].Frontiers in Plant Science,2016,7:1077.doi:10.3389/fpls.2016.1077.
[32] Huang Y F, Poland J A, Wight C P, Jackson E W, Tinker N A.Using Genotyping-By-Sequencing (GBS) for genomic discovery in cultivated oat[J].PLoS One,2014,9(7):102448.doi:10.1371/journal.pone.0102448.
[33] 刘龙龙, 张丽君, 马名川, 韩渊怀, 周建萍, 崔林. 山西省燕麦产业现状及发展趋势[J]. 山西农业大学学报(自然科学版), 2016, 36(12): 905-907, 912. doi:10.13842/j.cnki.issn1671-8151.2016.12.012.
Liu L L, Zhang L J, Ma M C, Han Y H, Zhou J P, Cui L. Current status and developing trend of oat industry in Shanxi[J]. Journal of Shanxi Agricultural University(Natural Science Edition), 2016,36(12):905-907, 912. doi:10.13842/j.cnki.issn1671-8151.2016.12.012.
[34] Schubert M, Lindgreen S, Orlando L.Adapter Removal v2:rapid adapter trimming,identification,and read merging[J].BMC Research Notes,2016,9(1):88. doi: 10.1186/s13104-016-1900-2.
[35] Catchen J, Hohenlohe P A, Bassham S, Amores A, Cresko W A.Stacks: an analysis tool set for population genomics[J].Molecular Ecology,2013,22(11):3124-3140.doi:10.1111/mec.12354.
[36] Owens D M, Keyse S M.Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases[J].Oncogene,2007,26(22):3203-3213.doi:10.1038/sj.onc.1210412.
[37] Aksaas A, Larsen A V, Rogne M, Rosendale K, Kvissel A K, Sk lhegg B S.G-patch domain and KOW motifs-containing protein,GPKOW;a nuclear RNA-binding protein regulated by protein kinase A[J].Journal of Molecular Signaling,2011,6(1):10. doi: 10.1186/1750-2187-6-10.
[38] Laloi C, Rayapuram N, Chartier Y, Rayapuram N, Chartier Y, Grienenberger J M, Bonnard J, Meyer Y.Identification and characterization of a mitochondrial thioredoxin system in plants[J].Proceedings of the National Academy of Sciences,2001,98(24):14144-14149. doi: 10.1073/pnas.241340898
[39] Wang Y P, Nishimura M T, Zhao T, Tang D Z.ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis[J].Plant Journal,2011,68(1):74-87.doi:10.1111/j.1365-313X.2011.04669.x.
[40] Colcombet J, Lopez-Obando M, Heurtevin L A, Martin K, Berthome R, Lurin C.Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles[J].RNA Biology,2013,10(9):1557-1575.doi:10.4161/rna.26128.
[41] Ganie S A, Pani D R, Mondal T K.Genome-wide analysis of DUF221 domaincontaining gene family in Oryza species and identification of its salinity stress-responsive members in rice[J].PLoS One,2017,12(8):e0182469.doi:10.1371/journal.pone.0182469.
[42] Banerjee A, Roychoudhury A. WRKY proteins: signaling and regulation of expression during abiotic stress responses[J]. The Scientific World Journal, 2015, 200: 807560. doi: 10.1155/2015/807560.
[43] Gu X B, Gao Z H, Yan Y C, Wang X Y, Qiao Y S, Chen Y H.RdreB1BI enhances drought tolerance by activating AQP-related genes in transgenic strawberry[J].Plant Physiology and Biochemistry,2017,119:33-42.doi:10.1016/j.plaphy.2017.08.013.
[44] Neale A D, Wahleithner J A, Lund M, Bonnett H T, Kelly A, Meeks-Wagner D R, Peacock W J, Dennis E S. Chitinase, beta-1,3-glucanase, osmotin, and extensin are expressed in tobacco explants during flower formation[J].The Plant Cell,1990,2(7):673-684.doi:10.1105/tpc.2.7.673.
[45] Even Y, Bennett J L, Sekulovic S, Rossi F V.NUP98-HOXA10hd-expanded hematopoietic stem cells efficiently reconstitute bone marrow of mismatched recipients and induce tolerance[J].Cell Transplantation,2011,20(7):1099-1108. doi: 10.3727/096368910X545068.
[46] Robledo D, Palaiokostas C, Bargelloni L, Mart nez P, Houston R.Applications of genotyping by sequencing in aquaculture breeding and genetics[J].Reviews in Aquaculture,2018,10(3):670-682. doi:org/10.1111/raq.12193.
[47] Crossa J, Pérez P,de los campos G,Muhuku G,Dreisigacker S,Magorokosho C.Genomic selection and prediction in plant breeding[J].Journal of Crop Improvement,2011,25(3):239-261. doi:org/10.1080/15427528.2011.558767.
[48] Makumburage G B, Richbourg H L, Latorre K D, Capps A, Chen C, Stapleton A E. Genotype to phenotype Maps: multiple input abiotic signals combine to produce growth effects via attenuating signaling interactions in maize[J].G3-Genes Genomes Genetics,2013,3(12):2195-2204.doi:10.1534/g3.113.008573.
[49] Mcnally K L, Naredo M E, Cairns J.SNP discovery at candidate genes for drought responsiveness in rice[M].Drought Frontiers in Rice:Crop Improvement for Increased Rainfed Production,2009:311-324. doi: 10.1142/9789814280013_0017.
[50] Homolka A, Eder T, Kopecky D, Berenyi M, Burg K, Fluch S.Allele discovery of ten candidate drought-response genes in Austrian oak using a systematically informatics approach based on 454 amplicon sequencing[J].BMC Research Notes,2012,5(1):175. doi: 10.1186/1756-0500-5-175.
[51] Cortés A J, Monserrate F A, Ram rez-Villegas J, Madri n S, Blair M W.Drought tolerance in wild plant populations: the case of common beans(Phaseolus vulgaris L.)[J].PLoS One,2013,8(5):e62898. doi:org/10.1371/journal.pone.0062898.
[52] Bhullar N K, Zhang Z Q, Wicker T, Keller B.Wheat gene bank accessions as a source of new alleles of the powdery mildew resistance gene Pm3: a large scale allele mining project[J].BMC Plant Biology,2010,10(88):1471-2229.doi:10.1186/1471-2229-10-88.
[53] Cattivelli L, Rizza F, Badeck F W, Mazzucotelli E, Mastrangelo A M, Francia E, Mar C, Tondelli A, Stanca A M. Drought tolerance improvement in crop plants: An integrated view from breeding to genomics[J].Field Crops Research,2008,105(1/2):1-14.doi:10.1016/j.fcr.2007.07.004.
[54] Abou-Elwafa S F.Identification of genes associated with drought tolerance in barley[J].Biologia Plantarum,2018,62(2):299-306.doi:10.1007/s10535-017-0765-0.
[55] Mir R R, Zaman-Allah M, Sreenivasulu N, Trethowan R, Varshney R K.Integrated genomics,physiology and breeding approaches for improving drought tolerance in crops[J].Theoretical and Applied Genetics,2012,125(4):625-645. doi:org/10.1007/s00122-012-1904-9.
[56] Tubersora R, Salvi S.Genomics-based approaches to improve drought tolerance of crops[J].Trends in Plant Science,2006,11(8):405-412.doi:org/10.1016/j.tplants. 2006.06.003.
[57] Lu G H, Wang X P, Liu J H, Yu K, Gao Y, Liu H Y, Wang C G, Wang W, Wang G A, Mao G F, Li B F, Qin J Y, Xia M, Zhou J L, Liu J M, Jiang S Q, Mo H A, Nagasawa N, Sivasankar S, Albertsen M C, Sakai H, Mazur B J, Lassner M W, Broglie R M. Application of T-DNA activation tagging to identify glutamate receptor-like genes that enhance drought tolerance in plants[J].Plant Cell Reports,2014,33(4):617-631. doi:10.1007/s00299-014-1586-7.
[58] Casaretto J A, El-Kereamy A, Zeng B, Stiegelmeyer S M, Chen X, Yong M B, Rothstein S J.Expression of OsMYB55 in maize activates stress-responsive genes and enhances heat and drought tolerance[J].BMC Genomics,2016,17(1):312. doi: 10.1186/s12864-016-2659-5.
[59] Webster H, Keeble G, Dell B, Fosu-Nyarko J, Mukai Y, Moolhuijzen P, Bellgard M, Jia J Z, Kong X Y, Feuillet C, Choulet F, Appels R, Int wheat genome sequencing consor. Genome-level identification of cell wall invertase genes in wheat for the study of drought tolerance[J].Functional Plant Biology,2012,39(7):569-579.doi:10.1071/FP12083.
[60] 段莹亮, 白洁. 硫氧还蛋白(Trx)及Trx80在免疫系统中的作用[J]. 免疫学杂志, 2009, 25(4): 483-486. doi:10.13431/j.cnki.immunol.j.20090129.
Duan Y L, Bai J. The role of thioredoxin(Trx)and Trx80 in the immune system[J]. Immunological Journal, 2009,25(4):483-486. doi:10.13431/j.cnki.immunol.j.20090129.
[61] Cha J Y, Kim J Y, Jung I J, Kim M R, Alam S S, Yun D J, Lee S Y, Kim M G, Kim W Y.NADPH-dependent thioredoxin reductase A (NTRA) confers elevated tolerance to oxidative stress and drought[J].Plant Physiology and Biochemistry,2014,80:184-191. doi:10.1016/j.plaphy.2014.04.008.
[62] Ozgur R, Uzilday B, Sekmen A H, Turkan I. Reactive oxygen species regulation and antioxidant defence in halophytes[J].Functional Plant Biology,2013,40(8/9, SI):832-847.doi:10.1071/FP12389.
[63] Zhai C Z, Zhao L, Yin L J, Chen M, Wang Q Y, Li L C, Xu Z S, Ma Y Z. Two wheat glutathione peroxidase genes whose products are located in chloroplasts improve salt and H2O2 tolerances in Arabidopsis[J].PLoS One,2013,8(10):e73989. doi:10.1371/journal.pone.0073989.
[64] 张帆, 蒋雷, 鞠丽萍, 金秀锋, 王轩, 张晓科, 王宏礼, 付晓洁. 一个普通小麦Trx超家族新基因TaNRX的克隆与抗旱相关标记开发[J]. 作物学报, 2014, 40(1): 29-36. doi:10.3724/SP.J.1006.2014.00029.
Zhang F, Jiang L, Ju L P, Jin X F, Wang X, Zhang X K, Wang H L, Fu X J. Cloning a novel gene TaNRX of Trx superfamily and developing its molecular markers related to drought resistance in common wheat[J]. Acta Agronomica Sinica, 2014,40(1):29-36. doi:10.3724/SP.J.1006.2014.00029.
[65] 张鹏钰, 袁珍, 王国瑞, 王同朝, 尹钧, 卫丽, 刘毓侠. 普通小麦Trx59基因的克隆及功能验证[J]. 华北农学报, 2017, 32(5): 1-6. doi:10.7668/hbnxb.2017.05.001.
Zhang P Y, Yuan Z, Wang G R, Wang T C, Yin J, Wei L, Liu Y X. Cloning and functional verification of Trx59 gene in common wheat[J]. Acta Agriculturae Boreali-Sinica, 2017,32(5):1-6. doi:10.7668/hbnxb.2017.05.001.