通过实时荧光定量PCR(Quantitative Real-time PCR,qRT-PCR)技术分析基因表达及调控,对于解析植物各种复杂的生物代谢途径和遗传性状及逆境胁迫生理响应具有重要的意义[1]。在利用qRT-PCR技术中,为了消除或降低不同组织间因RNA的提取质量、初始模板量或者反转录效率等所造成的偏差,必须选择适宜的内参基因进行校正和标准化[2]。选择稳定的内参基因也是保证其分析结果准确可靠的重要前提条件[3-4]。目前,Actin、GAPDH、18S rRNA、Ubiquitin等内参基因已经被广泛应用于qRT-PCR分析。然而,近年来大量的研究结果表明,任何一个内参基因不能够适用于所有条件下的试验研究[5-8]。所谓的持续稳定表达都只是限定在一定类型的细胞或试验因素作用下“有范围”的恒定[9-12]。据 Gutierrez等[13]报道,使用未经验证的内参基因,目标基因表达水平的定量会出现100倍的偏差。因此,在利用qRT-PCR进行定量分析之前,在不同试验材料或不同试验条件中,有必要验证、筛出稳定的内参基因,作为每个试验研究所适用的最佳内参基因稳定的定量因子。
本研究利用geNorm[14]和NormFinder[15]统计软件,比较分析了21个候选内参基因在大白菜-结球甘蓝易位系营养生长阶段的幼苗期、莲座期、包心期、结球期,及包心期IAA和TIBA处理的叶片和生殖生长阶段6个大小不同等级的花蕾的内参基因表达稳定性,旨在为精确分析大白菜-结球甘蓝易位系的基因表达提供保障,同时也为芸薹属植物不同发育时期及激素处理对内参基因的选择提供参考。
1 材料和方法
1.1 试验材料
河北省蔬菜种质创新与利用重点实验室创制的添加结球甘蓝(Brassica oleracea var. capitata)4号染色体片段、基因型纯合的大白菜(Brassica rapa ssp. pekinensis)易位系B3-29,于2015年8月8日播种于温室,进行育苗,9月12日进行田间定植。分别在幼苗期、莲座期、包心期和结球期的幼嫩叶片进行取材。并在包心期分别喷施生长素IAA(0.2,0.6,1.2 mmol/L)和生长素抑制剂TIBA(0.5,1.0,1.5 mmol/L),处理后12 h进行幼嫩叶片取材。2016年春季,待其抽薹开花取花蕾,分为6个阶段,花发育阶段1(花蕾横径介于1~2 mm)、花发育阶段2(花蕾横径介于2 ~3 mm)、花发育阶段3(花蕾横径大于3 mm,长度大于4 mm)、花发育阶段4(花蕾横径大于4 mm,长度大于5 mm)、花发育阶段5(花冠露出尚未伸展)、花发育阶段6(开花当天)。每个样品3个重复。所有样品液氮速冻后保存于-80 ℃冰箱备用。
1.2 实时荧光定量 PCR
本研究共检测了21个候选内参基因的表达稳定性:腺嘌呤转磷酸核糖基酶基因(Adeninephosphoribosyl transferase,Apr)、液泡膜内在蛋白基因(Tonop last intrinsic proteins,BcTIP41)、未知功能基因(Unknown protein,U34559)、延伸因子1基因(Elongationfactor1α,EF1α)、微管蛋白基因(Tubulin beta,TUB4)、亲环蛋白基因(Cyclophilin,CYP)、锌指蛋白基因(DnaJ-Like,DNAJ)、组蛋白基因(Histone h3,HIS)、α-微管蛋白基因(Alpha tubulin 5,TUA5)、假设蛋白基因(Hypothetical,UKN1)、互作蛋白基因(Ask-interacting protein 16,SKIP16)、网格衔接蛋白复合体基因(Clathrin adaptor complex,CAC)、肌动蛋白基因3个同源基因(ACTIN、ACTIN-1、ACTIN-2)、甘油醛-3-磷酸-脱氢酶基因(Glyc-eraldehyde-3-phosphate dehydrogenase,GAPDH)、泛素结合酶基因(Ubiquitin-conjugating enzyme,UBC30)、泛素连接酶基因(Poly ubiquitinenzyme,UBQ)、三角状五肽重复结构域基因(Pentatricopeptide repeat,PPR)、蛋白磷酸酶2A基因(Protein phosphatase 2A gene,PP2A)、苹果酸脱氢酶基因(Malate dehydrogenase,MDH)所有引物序列见表1。将样品的cDNA进行5个浓度梯度稀释,优化反应条件,确定检测样品的稀释浓度。qRT-PCR反应体系(20 μL):cDNA模板 2 μL,正反向引物(50 ng/μL)各0.5 μL,ddH2O 7 μL,THUN-DERBIRD SYBR qPCR Mix(QPS-201(-)TOYOBO公司)10 μL。反应程序:98 ℃预变性3 min;98 ℃变性30 s,58 ℃退火10 s,68 ℃延伸10 s,45个循环;之后65~95 ℃熔解曲线分析。每个样品均3次重复。试验使用罗氏LightCycler96实时荧光定量PCR仪定量分析系统进行数据采集。本试验所用试剂IAA和TIBA购于SIGMA公司,纯度大于98%。用RNA提取试剂盒ESAYspin Plus(北京艾德莱公司)分别提取样品总RNA。反转录根据ReverTra Ace qPCR RT Master Mix 试剂盒(东洋纺生物科技有限公司)说明进行cDNA合成。
表1 内参引物序列
Tab.1 Primers used for candidate reference genes
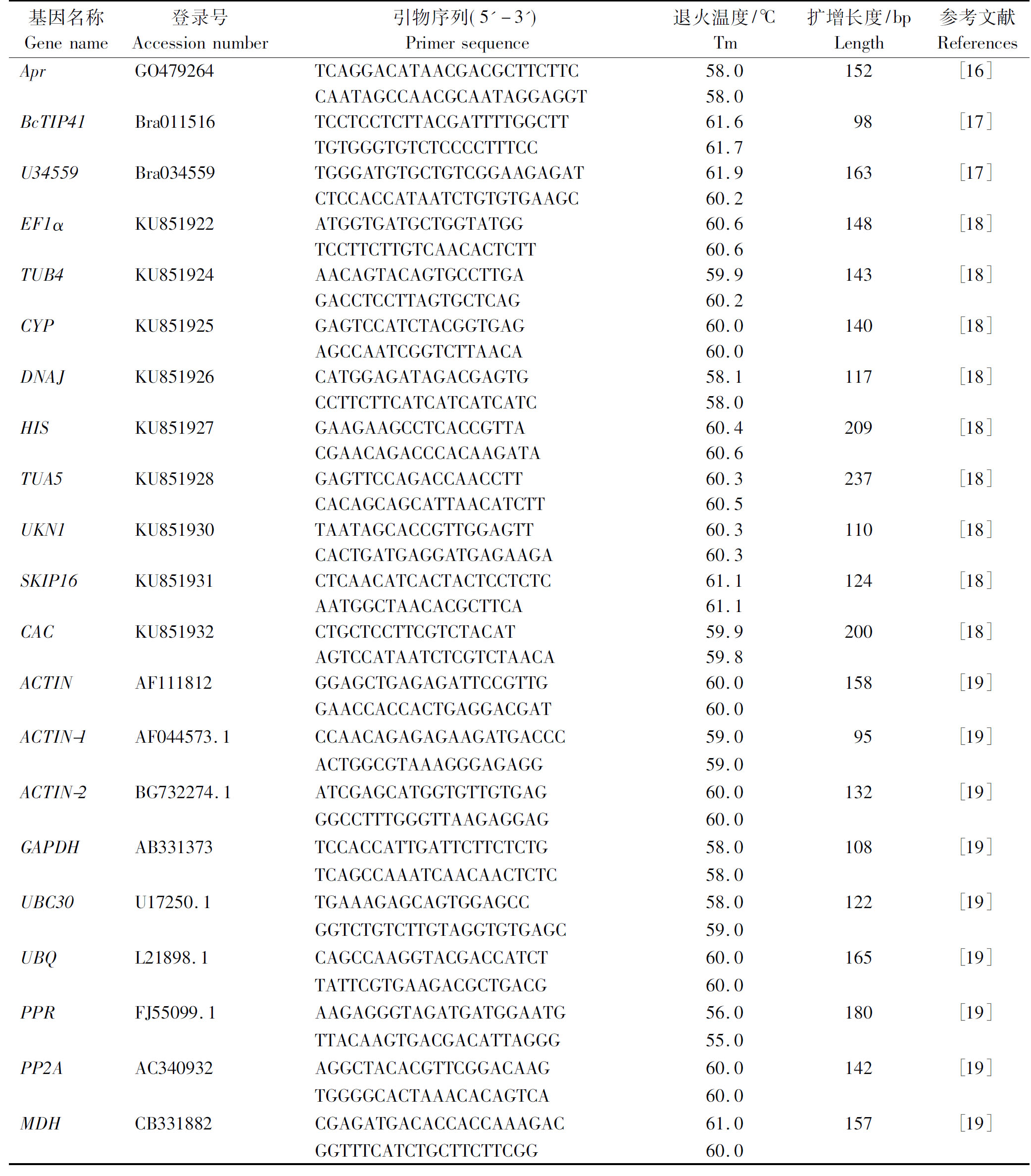
基因名称Gene name登录号Accession number引物序列(5'-3')Primer sequence退火温度/℃Tm扩增长度/bpLength 参考文献ReferencesApr GO479264TCAGGACATAACGACGCTTCTTC58.0152[16]CAATAGCCAACGCAATAGGAG-GT58.0BcTIP41Bra011516TCCTCCTCTTACGATTTTGGCTT61.698[17]TGTGGGTGTCTCCCCTTTCC61.7U34559 Bra034559TGGGATGTGCTGTCG-GAAGAGAT61.9163[17]CTCCACCATAATCTGTGTGAAGC60.2EF1αKU851922ATGGTGATGCTGGTATGG60.6148[18]TCCTTCTTGTCAACACTCTT60.6TUB4 KU851924AACAGTACAGTGCCTTGA59.9143[18]GACCTCCTTAGTGCTCAG60.2CYP KU851925GAGTCCATCTACGGTGAG60.0140[18]AGCCAATCGGTCTTAACA60.0DNAJ KU851926CATGGAGATAGACGAGTG58.1117[18]CCTTCTTCATCATCATCATC58.0HIS KU851927GAAGAAGCCTCACCGTTA60.4209[18]CGAACAGACCCACAAGATA60.6TUA5 KU851928GAGTTCCAGACCAACCTT60.3237[18]CACAGCAGCATTAACATCTT60.5UKN1 KU851930TAATAGCACCGTTGGAGTT60.3110[18]CACTGATGAGGATGAGAAGA60.3SKIP16 KU851931CTCAACATCACTACTCCTCTC61.1124[18]AATGGCTAACACGCTTCA61.1CAC KU851932CTGCTCCTTCGTCTACAT59.9200[18]AGTCCATAATCTCGTCTAACA59.8ACTIN AF111812GGAGCTGAGAGATTCCGTTG 60.0158[19]GAACCACCACTGAGGACGAT60.0ACTIN-1AF044573.1CCAACAGAGAGAAGATGACCC59.095[19]ACTGGCGTAAAGGGAGAGG59.0ACTIN-2 BG732274.1ATCGAGCATGGTGTTGTGAG60.0132[19]GGCCTTTGGGTTAAGAGGAG60.0GAPDH AB331373TCCACCATTGATTCTTCTCTG58.0108[19]TCAGCCAAATCAACAACTCTC58.0UBC30U17250.1TGAAAGAGCAGTGGAGCC58.0122[19]GGTCTGTCTTGTAGGTGTGAGC59.0UBQ L21898.1CAGCCAAGGTACGACCATCT60.0165[19]TATTCGTGAAGACGCTGACG60.0PPR FJ55099.1AAGAGGGTAGATGATGGAATG56.0180[19]TTACAAGTGACGACATTAGGG55.0PP2AAC340932AGGCTACACGTTCGGACAAG60.0142[19]TGGGGCACTAAACACAGTCA60.0MDH CB331882CGAGATGACACCACCAAAGAC 61.0157[19]GGTTTCATCTGCTTCTTCGG60.0
1.3 数据处理和分析
采用常用的内参分析软件geNorm(ver. 3.5)和NormFinder(ver. 0.953)进行数据处理和分析。根据qRT-PCR结果中Ct值,利用公式Q=E-ΔCt,计算出各基因的相对表达量Q[20]。利用geNorm软件对各候选内参基因的相对表达量Q进行表达稳定性统计学分析。其中,M值越小,稳定性越高。利用标准化因子的配对差异分析(V),确定合适的内参基因数目。该程序默认V值为0.15,若Vn/n+1>0.15,则引入第n+1个内参基因[14]。NormFinder软件通过方差分析得出基因的表达稳定值M,直接评价基因的稳定性[15]。
2 结果与分析
2.1 候选内参基因表达水平
对于21个候选内参基因,通过计算Ct值来比较基因的表达丰度,结果表明,试验涉及的21个内参基因在大白菜-结球甘蓝易位系不同类型材料中的平均Ct值为17.28~31.98(表2,3),并且大部分Ct值都集中在20.00~25.00,可用于后续试验。其中,GAPDH和MDH表达量较高,Ct值分别在19.00和20.00左右,PPR表达量较低,Ct值在30左右。从所有材料中的21个内参基因平均Ct值的箱式图中可以看出(图1),GAPDH和SKIP16基因的平均Ct值的最大值与最小值浮动范围较大。
表2 实时定量PCR分析中21个候选基因在营养生长阶段和激素处理的平均Ct值
Tab.2 Mean Ct values of 21 reference genes across vegetative growth stage and hormone treatment experimental in qRT-PCR
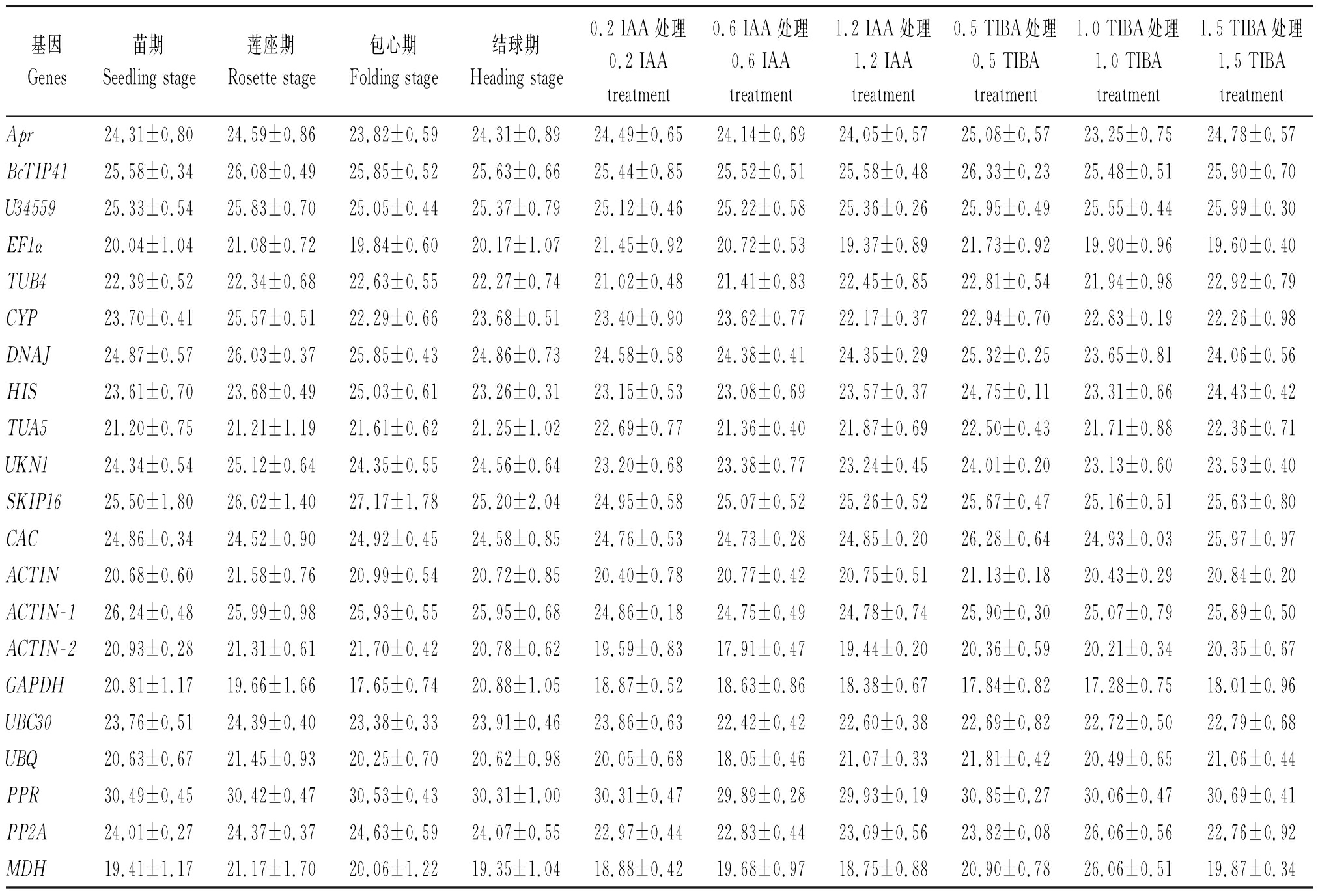
基因Genes苗期Seedling stage莲座期Rosette stage包心期Folding stage结球期Heading stage0.2 IAA 处理0.2 IAAtreatment0.6 IAA 处理 0.6 IAAtreatment1.2 IAA 处理1.2 IAAtreatment0.5 TIBA处理0.5 TIBAtreatment1.0 TIBA处理1.0 TIBAtreatment1.5 TIBA处理1.5 TIBAtreatmentApr24.31±0.8024.59±0.8623.82±0.5924.31±0.8924.49±0.6524.14±0.6924.05±0.5725.08±0.5723.25±0.7524.78±0.57BcTIP4125.58±0.3426.08±0.4925.85±0.5225.63±0.6625.44±0.8525.52±0.5125.58±0.4826.33±0.2325.48±0.5125.90±0.70U3455925.33±0.5425.83±0.7025.05±0.4425.37±0.7925.12±0.4625.22±0.5825.36±0.2625.95±0.4925.55±0.4425.99±0.30EF1α20.04±1.0421.08±0.7219.84±0.6020.17±1.0721.45±0.9220.72±0.5319.37±0.8921.73±0.9219.90±0.9619.60±0.40TUB422.39±0.5222.34±0.6822.63±0.5522.27±0.7421.02±0.4821.41±0.8322.45±0.8522.81±0.5421.94±0.9822.92±0.79CYP23.70±0.4125.57±0.5122.29±0.6623.68±0.5123.40±0.9023.62±0.7722.17±0.3722.94±0.7022.83±0.1922.26±0.98DNAJ24.87±0.5726.03±0.3725.85±0.4324.86±0.7324.58±0.5824.38±0.4124.35±0.2925.32±0.2523.65±0.8124.06±0.56HIS23.61±0.7023.68±0.4925.03±0.6123.26±0.3123.15±0.5323.08±0.6923.57±0.3724.75±0.1123.31±0.6624.43±0.42TUA521.20±0.7521.21±1.1921.61±0.6221.25±1.0222.69±0.7721.36±0.4021.87±0.6922.50±0.4321.71±0.8822.36±0.71UKN124.34±0.5425.12±0.6424.35±0.5524.56±0.6423.20±0.6823.38±0.7723.24±0.4524.01±0.2023.13±0.6023.53±0.40SKIP1625.50±1.8026.02±1.4027.17±1.7825.20±2.0424.95±0.5825.07±0.5225.26±0.5225.67±0.4725.16±0.5125.63±0.80CAC24.86±0.3424.52±0.9024.92±0.4524.58±0.8524.76±0.5324.73±0.2824.85±0.2026.28±0.6424.93±0.0325.97±0.97ACTIN20.68±0.6021.58±0.7620.99±0.5420.72±0.8520.40±0.7820.77±0.4220.75±0.5121.13±0.1820.43±0.2920.84±0.20ACTIN-126.24±0.4825.99±0.9825.93±0.5525.95±0.6824.86±0.1824.75±0.4924.78±0.7425.90±0.3025.07±0.7925.89±0.50ACTIN-220.93±0.2821.31±0.6121.70±0.4220.78±0.6219.59±0.8317.91±0.4719.44±0.2020.36±0.5920.21±0.3420.35±0.67GAPDH20.81±1.1719.66±1.6617.65±0.7420.88±1.0518.87±0.5218.63±0.8618.38±0.6717.84±0.8217.28±0.7518.01±0.96UBC3023.76±0.5124.39±0.4023.38±0.3323.91±0.4623.86±0.6322.42±0.4222.60±0.3822.69±0.8222.72±0.5022.79±0.68UBQ20.63±0.6721.45±0.9320.25±0.7020.62±0.9820.05±0.6818.05±0.4621.07±0.3321.81±0.4220.49±0.6521.06±0.44PPR30.49±0.4530.42±0.4730.53±0.4330.31±1.0030.31±0.4729.89±0.2829.93±0.1930.85±0.2730.06±0.4730.69±0.41PP2A24.01±0.2724.37±0.3724.63±0.5924.07±0.5522.97±0.4422.83±0.4423.09±0.5623.82±0.0826.06±0.5622.76±0.92MDH19.41±1.1721.17±1.7020.06±1.2219.35±1.0418.88±0.4219.68±0.9718.75±0.8820.90±0.7826.06±0.5119.87±0.34
注:IAA和TIBA处理浓度单位为mmol/L。
Note:The concentration unit of IAA and TIBA treatment is mmol/L.
2.2 候选内参基因表达稳定性分析
2.2.1 geNorm 分析 通过geNorm软件计算在大白菜-结球甘蓝易位系不同样品中各候选内参基因表达的M值,进行稳定性分析。结果显示(表4),在大白菜-结球甘蓝易位系营养生长阶段和生殖生长的不同时期或是在IAA和TIBA激素处理后,各候选内参基因表达稳定性有明显差异。在大白菜-结球甘蓝易位系营养生长阶段幼苗期、莲座期、包心期和结球期UKN1和TUB4基因表达较为稳定。在大白菜-结球甘蓝易位系包心期用不同浓度生长素IAA处理后,均表现出最稳定的内参基因为BcTIP41和ACTIN。用不同浓度生长素抑制剂TIBA处理后,最稳定的内参基因均为UKN1和BcTIP41。生殖生长时期花发育阶段的6个等级大小花蕾中最稳定内参基因为DNAJ和ACTIN,其次为PP2A。在总样品分析中认为,内参基因为BcTIP41和U34559较为稳定,其次为ACTIN,而内参基因SKIP16、GAPDH和MDH表现不稳定。
表3 实时荧光定量PCR分析中21个候选基因在花蕾发育阶段的平均Ct值
Tab.3 Mean Ct values of 21 reference genes across floral development stage experimental in qRT-PCR
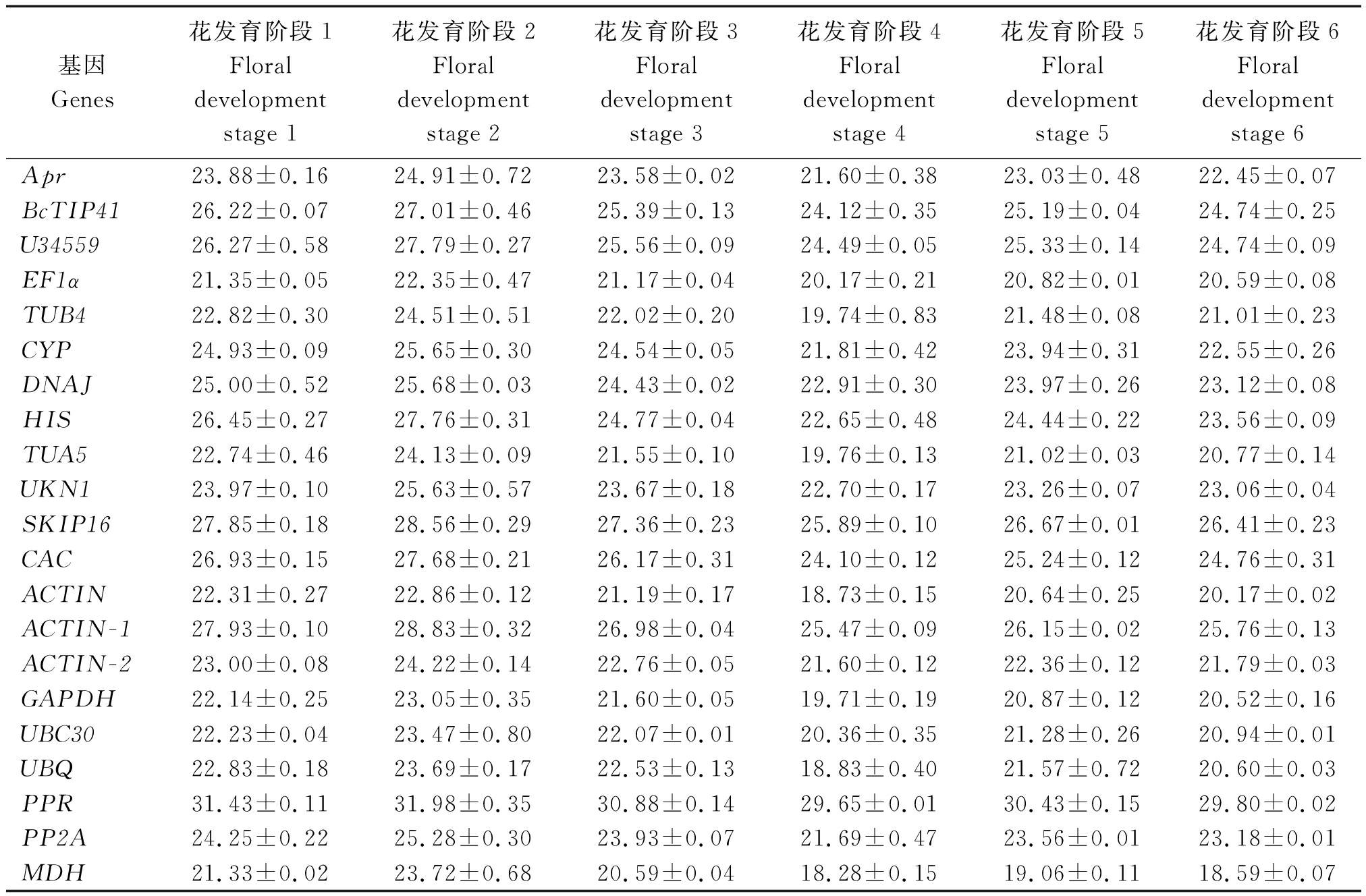
基因Genes花发育阶段1Floral developmentstage 1花发育阶段2Floraldevelopmentstage 2花发育阶段3Floraldevelopmentstage 3花发育阶段4Floraldevelopmentstage 4花发育阶段5Floraldevelopmentstage 5花发育阶段6Floraldevelopmentstage 6Apr23.88±0.1624.91±0.7223.58±0.0221.60±0.3823.03±0.4822.45±0.07BcTIP4126.22±0.0727.01±0.4625.39±0.1324.12±0.3525.19±0.0424.74±0.25U3455926.27±0.5827.79±0.2725.56±0.0924.49±0.0525.33±0.1424.74±0.09EF1α21.35±0.0522.35±0.4721.17±0.0420.17±0.2120.82±0.0120.59±0.08TUB422.82±0.3024.51±0.5122.02±0.2019.74±0.8321.48±0.0821.01±0.23CYP24.93±0.0925.65±0.3024.54±0.0521.81±0.4223.94±0.3122.55±0.26DNAJ25.00±0.5225.68±0.0324.43±0.0222.91±0.3023.97±0.2623.12±0.08HIS26.45±0.2727.76±0.3124.77±0.0422.65±0.4824.44±0.2223.56±0.09TUA522.74±0.4624.13±0.0921.55±0.1019.76±0.1321.02±0.0320.77±0.14UKN123.97±0.1025.63±0.5723.67±0.1822.70±0.1723.26±0.0723.06±0.04SKIP1627.85±0.1828.56±0.2927.36±0.2325.89±0.1026.67±0.0126.41±0.23CAC26.93±0.1527.68±0.2126.17±0.3124.10±0.1225.24±0.1224.76±0.31ACTIN22.31±0.2722.86±0.1221.19±0.1718.73±0.1520.64±0.2520.17±0.02ACTIN-127.93±0.1028.83±0.3226.98±0.0425.47±0.0926.15±0.0225.76±0.13ACTIN-223.00±0.0824.22±0.1422.76±0.0521.60±0.1222.36±0.1221.79±0.03GAPDH22.14±0.2523.05±0.3521.60±0.0519.71±0.1920.87±0.1220.52±0.16UBC3022.23±0.0423.47±0.8022.07±0.0120.36±0.3521.28±0.2620.94±0.01UBQ22.83±0.1823.69±0.1722.53±0.1318.83±0.4021.57±0.7220.60±0.03PPR31.43±0.1131.98±0.3530.88±0.1429.65±0.0130.43±0.1529.80±0.02PP2A24.25±0.2225.28±0.3023.93±0.0721.69±0.4723.56±0.0123.18±0.01MDH21.33±0.0223.72±0.6820.59±0.0418.28±0.1519.06±0.1118.59±0.07
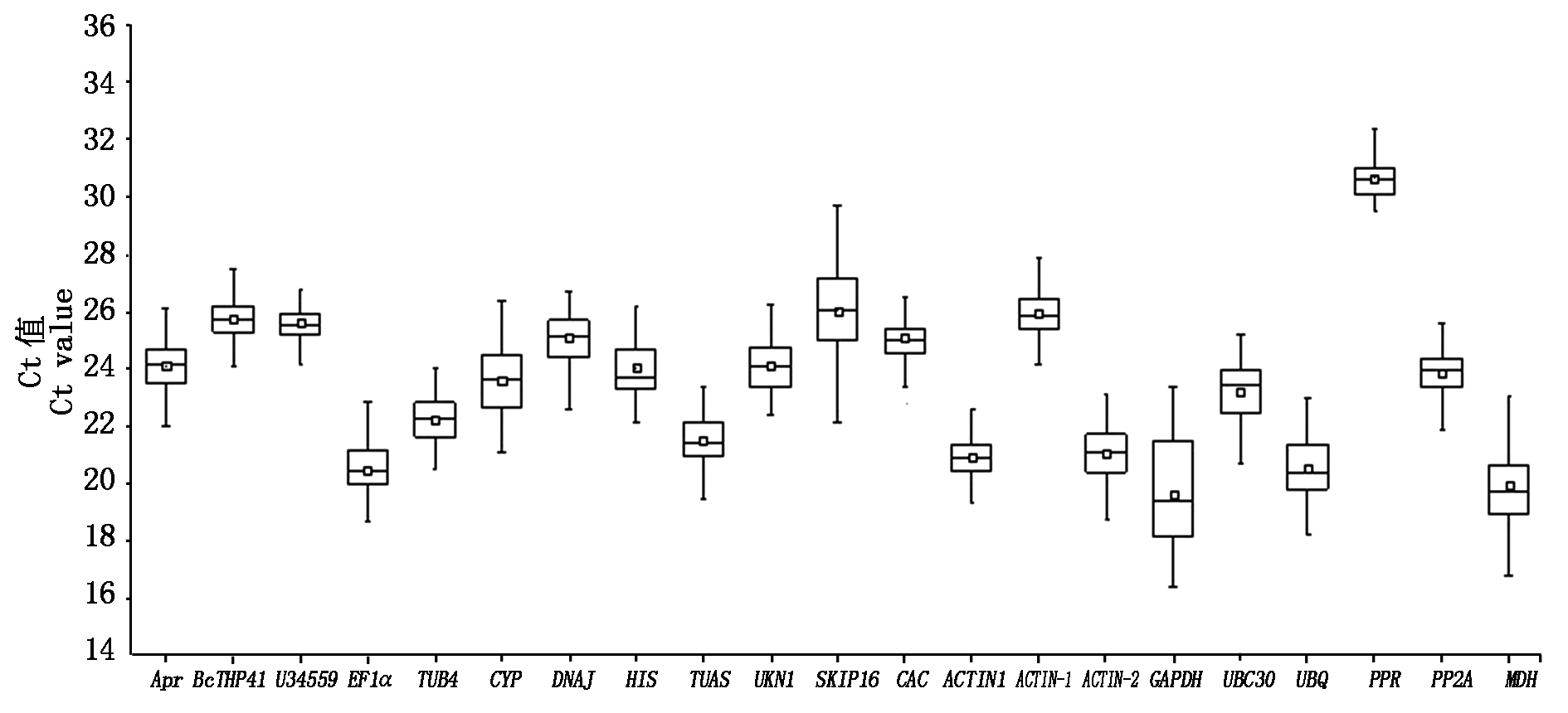
箱式图中线表示Ct值的中位数。内部小方块代表均值。上面四分之三、下面四分之一位数;上缘值和下缘值表示Ct数据中的极大值和极小值。 The line across the box depicts median. The inside box depicts mean. The outside box is determined by the 25th and 75th percentiles;The whiskers indicate the maximum and minimum values.
图1 实时荧光定量PCR分析中21个候选基因表达量
Fig.1 Expression levels of 21 candidate reference genes from all analyzed samples in qRT-PCR
为了获得更加准确可靠的RT-qPCR分析结果,通常会使用2个或更多内参基因进行校正。利用geNorm软件进行标准化因子的配对差异分析(V),确定各组合样品中最佳内参基因数目,由图2结果可知,对总样品分析中,V3/4的值是0.12,小于界限值0.15。因此,在总样品分析中,一组最优的稳定的内参基因至少需要3个,分别为:BcTIP41、U34559和ACTIN。同时花发育阶段组合V3/4的值是0.14,小于界限值0.15,表明在花发育阶段也需选用3个最稳定的内参基因进行标准化,分别为:DNAJ、ACTIN和PP2A。营养生长阶段和激素处理后材料的V2/3的值均小于界限值0.15,故只需选用2个最稳定的内参基因进行标准化。在营养生长阶段(从幼苗到结球)最稳定的内参基因为UKN1和TUB4。在包心期用不同浓度生长素IAA处理后,内参基因BcTIP41和ACTIN最为稳定,用不同浓度生长素抑制剂TIBA处理后,内参基因UKN1和BcTIP41最为稳定。
2.2.2 NormFinder软件分析 为了进一步验证geNorm分析的结果,笔者也使用了NormFinder软件进行稳定性分析。2个软件推荐的最稳定和最不稳定的基因比较一致(表4),如:在2个软件的分析显示,在总样品、营养生长阶段的幼苗期到结球期、生长素抑制剂TIBA处理后推荐的最为稳定的和最不稳定内参基因相一致。而在生长素IAA处理后最稳定内参基因BcTIP41的排名在2个软件存在一些差别,在geNorm软件中与ACTIN基因的M值相同排在第1位,而NormFinder软件分析却排在第4位,但是BcTIP41的M值与排在第2位基因仅相差0.06。在花发育阶段也出现了类似的现象,但是M值之间差距较小。
表4 geNorm和NormFinder分析21个内参基因的表达稳定值(M)
Tab.4 Gene expression stability of 21 candidate genes as predicted by geNorm and NormFinder
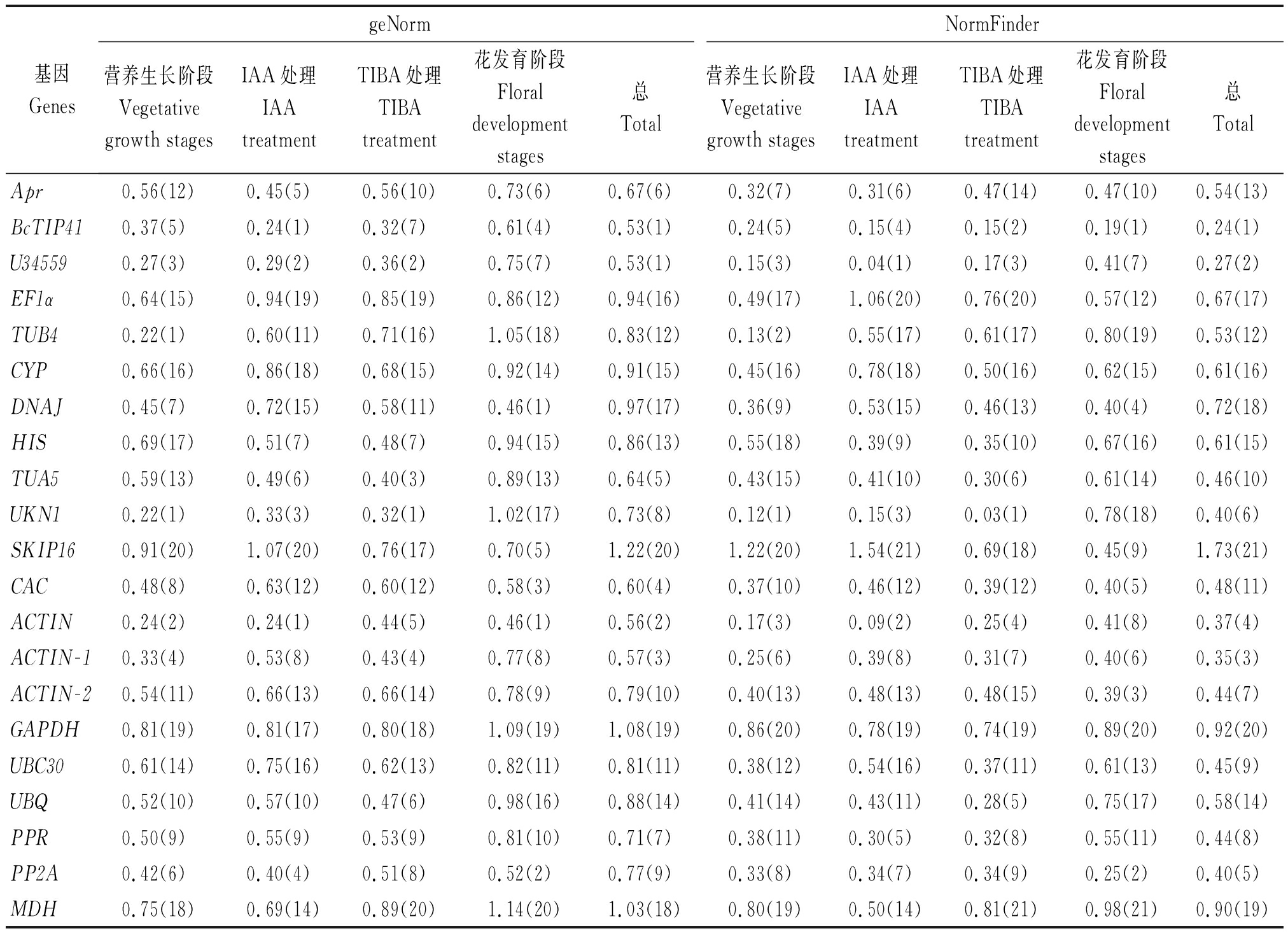
基因GenesgeNormNormFinder营养生长阶段Vegetativegrowth stagesIAA处理IAAtreatmentTIBA处理TIBAtreatment花发育阶段Floraldevelopmentstages总Total营养生长阶段Vegetativegrowth stagesIAA处理IAAtreatmentTIBA处理TIBAtreatment花发育阶段Floraldevelopmentstages总TotalApr0.56(12)0.45(5)0.56(10)0.73(6)0.67(6)0.32(7)0.31(6)0.47(14)0.47(10)0.54(13)BcTIP410.37(5)0.24(1)0.32(7)0.61(4)0.53(1)0.24(5)0.15(4)0.15(2)0.19(1)0.24(1)U345590.27(3)0.29(2)0.36(2)0.75(7)0.53(1)0.15(3)0.04(1)0.17(3)0.41(7)0.27(2)EF1α0.64(15)0.94(19)0.85(19)0.86(12)0.94(16)0.49(17)1.06(20)0.76(20)0.57(12)0.67(17)TUB40.22(1)0.60(11)0.71(16)1.05(18)0.83(12)0.13(2)0.55(17)0.61(17)0.80(19)0.53(12)CYP0.66(16)0.86(18)0.68(15)0.92(14)0.91(15)0.45(16)0.78(18)0.50(16)0.62(15)0.61(16)DNAJ0.45(7)0.72(15)0.58(11)0.46(1)0.97(17)0.36(9)0.53(15)0.46(13)0.40(4)0.72(18)HIS0.69(17)0.51(7)0.48(7)0.94(15)0.86(13)0.55(18)0.39(9)0.35(10)0.67(16)0.61(15)TUA50.59(13)0.49(6)0.40(3)0.89(13)0.64(5)0.43(15)0.41(10)0.30(6)0.61(14)0.46(10)UKN10.22(1)0.33(3)0.32(1)1.02(17)0.73(8)0.12(1)0.15(3)0.03(1)0.78(18)0.40(6)SKIP160.91(20)1.07(20)0.76(17)0.70(5)1.22(20)1.22(20)1.54(21)0.69(18)0.45(9)1.73(21)CAC0.48(8)0.63(12)0.60(12)0.58(3)0.60(4)0.37(10)0.46(12)0.39(12)0.40(5)0.48(11)ACTIN0.24(2)0.24(1)0.44(5)0.46(1)0.56(2)0.17(3)0.09(2)0.25(4)0.41(8)0.37(4)ACTIN-10.33(4)0.53(8)0.43(4)0.77(8)0.57(3)0.25(6)0.39(8)0.31(7)0.40(6)0.35(3)ACTIN-20.54(11)0.66(13)0.66(14)0.78(9)0.79(10)0.40(13)0.48(13)0.48(15)0.39(3)0.44(7)GAPDH0.81(19)0.81(17)0.80(18)1.09(19)1.08(19)0.86(20)0.78(19)0.74(19)0.89(20)0.92(20)UBC300.61(14)0.75(16)0.62(13)0.82(11)0.81(11)0.38(12)0.54(16)0.37(11)0.61(13)0.45(9)UBQ0.52(10)0.57(10)0.47(6)0.98(16)0.88(14)0.41(14)0.43(11)0.28(5)0.75(17)0.58(14)PPR0.50(9)0.55(9)0.53(9)0.81(10)0.71(7)0.38(11)0.30(5)0.32(8)0.55(11)0.44(8)PP2A0.42(6)0.40(4)0.51(8)0.52(2)0.77(9)0.33(8)0.34(7)0.34(9)0.25(2)0.40(5)MDH0.75(18)0.69(14)0.89(20)1.14(20)1.03(18)0.80(19)0.50(14)0.81(21)0.98(21)0.90(19)
注:括号中为稳定性排序。
Note:Stability ranking in parenthese.
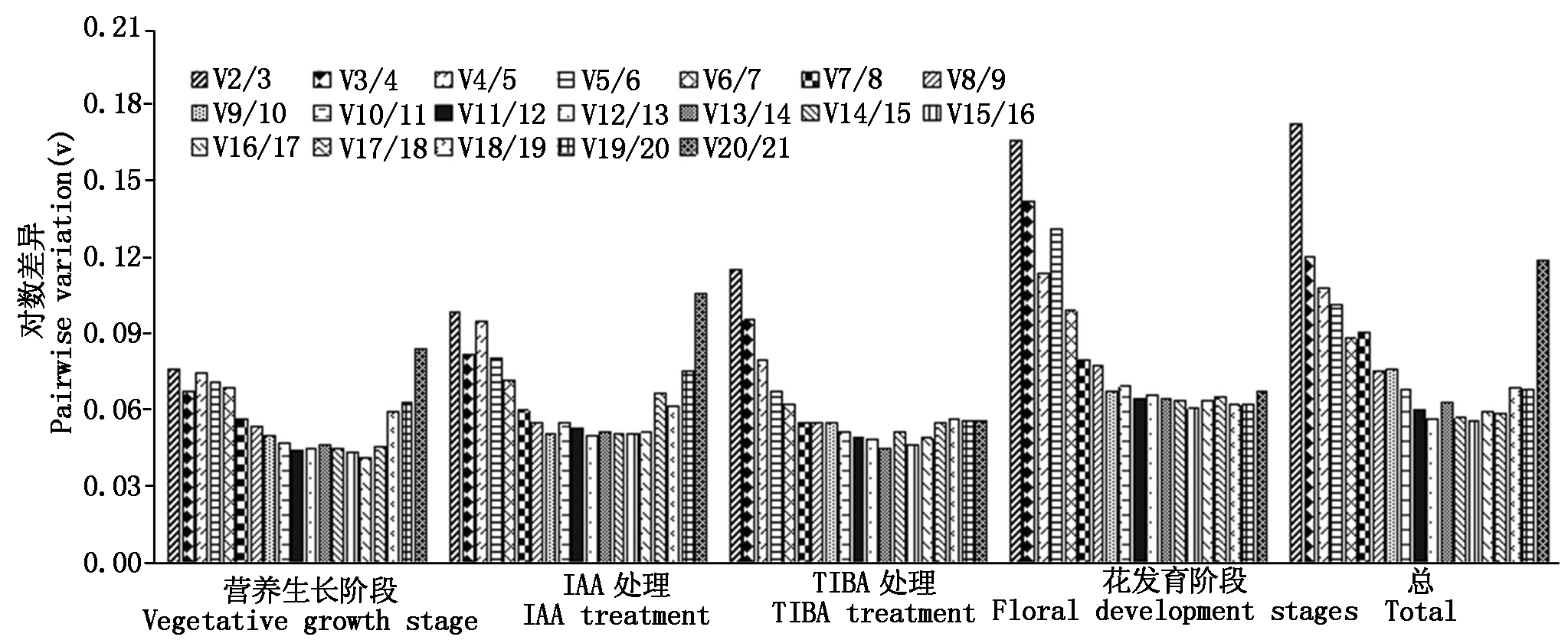
当Vn/n+l<0.15时,优化的内标基因个数为N。 When Vn/n+1 < 0.15 then the optimal number of reference genes is N.
图2 候选内标基因的对数差异检测
Fig.2 Pairwise variation(V)measure of the candidate reference genes
3 讨论与结论
目前,在多数植物中都鉴定出了各自在不同的试验条件下表达最为稳定的内参基因,包括大豆(Glycine max)、小拟南芥(Arabidopsis pumila)、野生条斑紫菜(Pyropia yezoensis)、牡丹(Paeonia suffruticosa)、菊花(Chrysanthemum lavandulifolium)、鼠尾草(Salvia hispanica)、茄子(Solanum melongena L.)[21-27]。研究结果表明,内参基因GAPDH在葡萄(Vitis amurensis)和荔枝(Litchi chinensis Sonn.)中表达均较稳定,而在黑麦草(Lolium perenne)、芥菜(Brassica juncea)、柳枝稷(Panicum virgatum L.)中的表达稳定性则较差 [28-32]。在菊花中能够稳定表达的SKIP16,在大白菜花的发育中则稳定性较低[18]。李晗等[33]报道,在羽衣甘蓝的不同组织中GAPDH为最不稳定内参基因。本研究中,GAPDH和SKIP16基因在大白菜-结球甘蓝易位系营养生长时期叶球发育过程中及IAA和TIBA处理后都不能稳定表达。表明在不同物种中,同一内参基因的表达稳定性各不相同。
本研究大白菜-结球甘蓝易位系不同发育阶段、不同组织部位以及不同激素处理条件下,21个候选内参基因的表达稳定性也各不相同。Zhang等[34]对麻风树(Jatropha curcas)内参基因分析,认为营养生长阶段ACT和TUB8内参基因最稳定,在生殖生长阶段内参基因GAPDH和EF-1α最为稳定。吝月爱[35]在玉米的不同组织中认为,内参基因TUB、UBQ9和EF1lα是茎中最稳定;EF1α、UBQ9和GAPDH是叶片中最稳定;在激素处理条件下,EF1α、TUB和GAPDH在脱落酸处理条件下最稳定;ACT2和TUB是赤霉素处理条件下最稳定。刘洪峰等[24]在牡丹内参基因筛选时,认为不同组织(茎、叶片、花萼、花瓣)中内参基因AMPDS和PUF1639最为稳定,在不同发育时期的种子中认为RPS9和PUF1639基因最为稳定。进一步证明了同一植物中的不同发育阶段、不同组织部位及不同处理条件下,内参基因的稳定性不尽相同。
Cheng等[18]对13个内参基因在不结球大白菜花的不同发育阶段及不同花器官组织部位中进行内参基因筛选,其中,DNAJ、UKN1和PP2A被确定为最稳定的内参基因。本研究对大白菜-结球甘蓝易位系花发育阶段的6个等级大小的花蕾进行21个内参基因筛选发现,最稳定的内参基因为DNAJ、ACTIN和PP2A,与Cheng等[18]研究结果相似。本研究选择的内参基因在包含了Cheng等[18]研究的12个内参基因基础上,又增加了9个内参基因进行筛选,共21个内参基因,其中,DNAJ和PP2A在不结球大白菜、大白菜-结球甘蓝易位系的花器官中都表现了高稳定性,表明该研究不仅为大白菜-结球甘蓝易位系基因表达量的精准分析提供了必要保障,同时也为芸薹属其他植物内参基因的选择提供了参考。
在大白菜-结球甘蓝易位系营养生长阶段(从幼苗到结球)内参基因UKN1和TUB4表达最为稳定。生长素IAA处理后,最稳定的内参基因为BcTIP41和ACTIN,生长素抑制剂TIBA处理后,最稳定的内参基因为UKN1和BcTIP41。在花发育阶段6个大小等级花蕾中DNAJ、ACTIN和PP2A基因表达稳定。内参基因GAPDH和MDH在大白菜-结球甘蓝易位系的营养生长阶段、激素IAA和TIBA处理及花发育阶段表达均不稳定。
[1] Mascia T,Santovito E,Gallitelli D,et al. Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants[J]. Molecular Plant Pathology,2010,11(6):805-816.
[2] Derveaux S,Vandesompele J,Hellemans J. How to do successful gene expression analysis using Real-time PCR[J]. Methods,2010,50(4):227-230.
[3] Kozera B,Rapacz M. Reference genes in Real-time PCR[J]. Journal of Applied Genetics,2013,54(4):391-406.
[4] Vanguilder H D,Vrana K E,Freeman W M. Twenty-five years of quantitative PCR for gene expression analysis[J]. BioTechniques,2008,44(5):619-626.
[5] Dong M,Zhang X,Chi X,et al. The validity of a reference gene is highly dependent on the experimental conditions in green alga Ulva linza[J]. Current Genetics,2012,58(1):13-20.
[6] 林榕燕,钟淮钦,黄敏玲,等.杂交兰实时荧光定量PCR内参基因的筛选[J].中国细胞生物学学报,2018,40(3):381-389.
[7] Zhou X H,Liu J,Zhuang Y. Selection of appropriate reference genes in eggplant for quantitative gene expression studies under Different experimental conditions[J]. Scientia Horticulturae,2014,176(2):200-207.
[8] Chen H,Yang Z Q,Hu Y,et al. Reference genes selection for quantitative gene expression studies in Pinus massoniana L.[J]. Trees,2016,30(3):685-696.
[9] Long X Y,Wang J R,Ouellet T,et al. Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat[J]. Plant Molecular Biology,2010,74(3):307-311.
[10] Hua W,Zhu J,Shang Y,et al. Identification of suitable reference genes for barley gene expression under abiotic stresses and hormonal treatments[J]. Plant Molecular Biology Reporter,2015,33(4):1002-1012.
[11] Xu Y,Hui L,Li X,et al. Systematic selection and validation of appropriate reference genes for gene expression studies by quantitative real-time PCR in pear[J]. Acta Physiologiae Plantarum,2015,37(2):40.
[12] Ma S,Niu H,Liu C,et al. Expression stabilities of candidate reference genes for RT-qPCR under Different stress conditions in soybean[J]. PloS One,2013,8(10):e75271.
[13] Gutierrez L,Mauriat M,Gué nin S,et al. The lack of a systematic validation of reference genes:a serious pitfall undervalued in reverse transcription-polymerase chain reaction(RT-PCR)analysis in plants[J]. Plant Biotechnology Journal,2008,6(6):609-618.
[14] Vandesompele J,Preter K D,Pattyn F,et al. Accurate normalization of Real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes[J]. Genome Biology,2002,3(7):research0034.1.
[15] Andersen C L,Jensen J L,Orntoft T F. Normalization of Real-time quantitative reverse transcription-PCR data:A model-based variance estimation approach to identify genes suited for normalization,applied to bladder and colon cancer data sets[J]. Cancer Research,2004,64(15):5245-5250.
[16] Qi J,Yu S,Zhang F,et al. Reference gene selection for Real-time quantitative polymerase chain reaction of mRNA transcript levels in Chinese cabbage(Brassica rapa L. ssp. pekinensis)[J]. Plant Molecular Biology Reporter,2010,28(4):597-604.
[17] Xu X,Yang Z,Sun X,et al. Selection of reference genes for quantitative Real-time PCR during flower bud development in CMS7311 of heading Chinese cabbage(Brassica rapa L. ssp. pekinensis)[J]. Acta Physiologiae Plantarum,2014,36(3):809-814.
[18] Cheng W,Cui H M,Huang T H,et al. Identification and validation of reference genes for RT-qPCR analysis in non-heading Chinese cabbage flowers[J]. Frontiers in Plant Science,2016,7(651):811.
[19] Xiao D,Zhang N W,Zhao J J,et al. Validation of reference genes for Real-time quantitative PCR normalisation in non-heading Chinese cabbage[J]. Functional Plant Biology,2012,39(4):342-350.
[20] 李钱峰,蒋美艳,于恒秀,等.水稻胚乳RNA定量RT-PCR分析中参照基因选择[J].扬州大学学报:农业与生命科学版,2008,29(2):61-66.
[21] 侯高杰.铝毒胁迫下大豆内参基因的筛选及相关基因表达分析[D].南宁:广西大学,2015.
[22] 郑丽洁,林 军,黄先忠.小拟南芥实时荧光定量PCR(qRT-PCR)内参基因的筛选[J].基因组学与应用生物学,2017(2):774-783.
[23] Kong F N,Cao M,Sun P P,et al. Selection of reference genes for gene expression normalization in Pyropia yezoensis using quantitative Real-time PCR[J]. Journal of Applied Phycology,2015,27(2):1003-1010.
[24] 刘洪峰,高乐旋,胡永红.牡丹不同发育阶段种子和花瓣组织实时荧光定量PCR中内参基因的挖掘与筛选[J].农业生物技术学报,2015,23(12):1639-1648.
[25] Shuai Q,Yang L,Wen X,et al. Reference gene selection for RT-qPCR analysis of flower development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium [J]. Frontiers in Plant Science,2016,7(287).
[26] Gopalam R,Rupwate S D,Tumaney A W. Selection and validation of appropriate reference genes for quantitative Real-time PCR analysis in Salvia hispanica[J]. PloS One,2017,12(11):e0186978.
[27] 庞强强,李植良,罗少波,等.高温胁迫下茄子qRT-PCR内参基因筛选及稳定性分析[J].园艺学报,2017,44(3):475-486.
[28] Peng F,James S,Niclas O,et al. An optimized grapevine RNA isolation procedure and statistical determination of reference genes for Real-time RT-PCR during berry development[J]. Bmc Plant Biology,2006,6(1):27.
[29] Zhong H Y,Chen J W,Li C Q,et al. Selection of reliable reference genes for expression studies by reverse transcription quantitative Real-time PCR in litchi under Different experimental conditions[J]. Plant Cell Reports,2011,30(4):641-653.
[30] Ruby C,Rehna A,Bisht N C. Evaluation of candidate reference genes for gene expression normalization in Brassica juncea using real time quantitative RT-PCR[J]. PloS One,2012,7(5):e36918.
[31] 严海东,蒋晓梅,张新全,等.非生物胁迫下多年生黑麦草qRT-PCR分析中内参基因的选择[J].农业生物技术学报,2014,22(12):1494-1501.
[32] 蒋晓梅,张新全,严海东,等.柳枝稷根组织实时定量PCR分析中内参基因的选择[J].农业生物技术学报,2014,22(1):55-63.
[33] 李 晗,李治龙,李晓屿,等.羽衣甘蓝不同组织及柱头发育实时荧光定量PCR内参基因的筛选[J].植物研究,2016,36(4):565-572.
[34] Zhang L, He L L, Fu Q T, et al. Selection of reliable reference genes for gene expression studies in the biofuel plant Jatropha curcas using Real-time quantitative PCR[J].International Journal of Molecular Sciences, 2013, 14(12):24338.
[35] 吝月爱.玉米在非生物胁迫和激素处理条件下实时荧光定量PCR内参基因的选择[D].雅安:四川农业大学,2012.