重金属污染已日益成为威胁人类健康的环境问题,而镉(Cd)因其移动性大、毒性持久而成为毒性最强的重金属元素之一[1]。长期暴露在镉污染环境中会导致人体健康受损,包括肺气肿、骨损伤(Itai-Itai)、癌症、心血管疾病和不可逆转的慢性肾功能衰竭[2-4]。
大豆因其含有丰富的营养物质,在世界上的消费越来越广泛[5-6]。但有研究报道,大豆较易吸收重金属[7]。最近的分子生物学研究发现,一些基因家族参与调控植物对Cd的吸收和转运[8-9],其中也包括了bHLH(Basic helix-loop-helix)转录因子家族[10]。
bHLH是普遍存在于真核生物中的转录因子家族,尤其是在哺乳动物中,其结构、功能和进化都得到了很好的分析[11-12]。近年来,有关bHLH在植物中的功能报道越来越多,主要在植物表皮分化、响应环境胁迫以及调控次生代谢调控中起着重要作用[13-15]。
前期对栽培大豆进行了Cd胁迫,并取其根系进行了转录组测序,对其差异基因进行分析发现,GmbHLH041基因在Cd胁迫后呈上调表达,推测该基因与大豆耐Cd性相关。本试验结合NCBI(National Center for Biotechnology Information)和Phytozome基因组数据库,并通过生物信息学软件,对大豆GmbHLH041基因进行进化树分析和同源序列比对,并通过荧光定量PCR分析Cd胁迫下大豆根系中GmbHLH041基因的表达情况,为进一步明确大豆GmbHLH041基因的功能以及作物在Cd胁迫后的应答机制提供理论基础。
1 材料和方法
1.1 大豆GmbHLH041基因获得及生物信息学分析
前期对Cd胁迫前后大豆根系进行了转录组测序,分析了差异基因的表达情况,结果发现,通过GmbHLH041基因(GenBank登录号为 XM_006600391)在Cd胁迫后表达量急剧上升。从NCBI数据库中获得GmbHLH041的序列,结合生物信息学分析软件 BioXM 2.6对大豆GmbHLH041蛋白理化性质进行分析。
大豆GmbHLH041蛋白进化树分析:首先使用Clustal X2对氨基酸序列进行比对分析,然后用 MEGA 6 软件中的邻接法(Neighbor-Joining,NJ)构建系统进化树,校验参数 Bootstrap 重复1 000 次。
保守序列多重比对:首先使用Clustal X2对氨基酸序列进行比对分析,导出的文件格式通过GeneDoc软件打开。
1.2 大豆幼苗培养和实时荧光定量PCR(qRT-PCR)分析
大豆种子用去离子水浸泡6 h,播种于营养土中,在光照培养箱内催芽。4 d后选取生长一致的幼苗移至1/2 Hoagland 营养液中培养,每2 d更换一次营养液。培养15 d后,用50 μmol/L CdSO4进行处理,取0,24 h的根系,液氮速冻于-80 ℃保存,用于目标基因诱导表达分析。
1.3 总 RNA 提取和 cDNA 合成
将50 μmol/L CdSO4 处理 0,24 h的根部组织用TRIzol 试剂进行提取,通过琼脂糖凝胶电泳检测总 RNA 的浓度和纯度。 采用TaKaRa反转录试剂盒反转RNA合成cDNA。
1.4 基因表达分析
根据NCBI中GmbHLH041基因序列设计引物,引物序列为F:5′-GAACACTTCTTCCTCCA-3′;R:5′-ACAACTTTCTCCTCTCAC-3′;以大豆tublin基因作为内参基因,tublin-F:5′-ATTCCCTTCCCTCGTCTG-3′;tublin-R:5′-CCTTGGTGCTCATCTTGC-3′,以cDNA为模板,按照 SYBR Premix Ex Taq(TaKaRa)反应体系进行定量 PCR 分析,以组成型表达的tublin 为内参,计算出目标基因的相对表达量,每个样品3次生物学重复。
2 结果与分析
2.1 GmbHLH041的克隆与序列分析
以大豆根系cDNA为模板,利用RT-PCR技术扩增出长度为1 629 bp的GmbHLH041的开放阅读框(Opening reading frame,ORF),编码542个氨基酸的蛋白质,GmbHLH041蛋白相对分子质量为61.17 ku,理论等电点为5.89,在Phytozome数据库比对发现,GmbHLH041基因位于大豆第17条染色体上。
用SOPMA对GmbHLH041蛋白氨基酸序列的二级结构预测结果显示,该氨基酸序列主要以无规则卷曲(39.48%)、α-螺旋(37.27%)和伸展链(16.61%)为主,含少量的β-转角(6.64%)(图1)。利用TMHMM 2.0 Server对GmbHLH041蛋白的跨膜结构域进行预测,发现该蛋白没有跨膜区段。
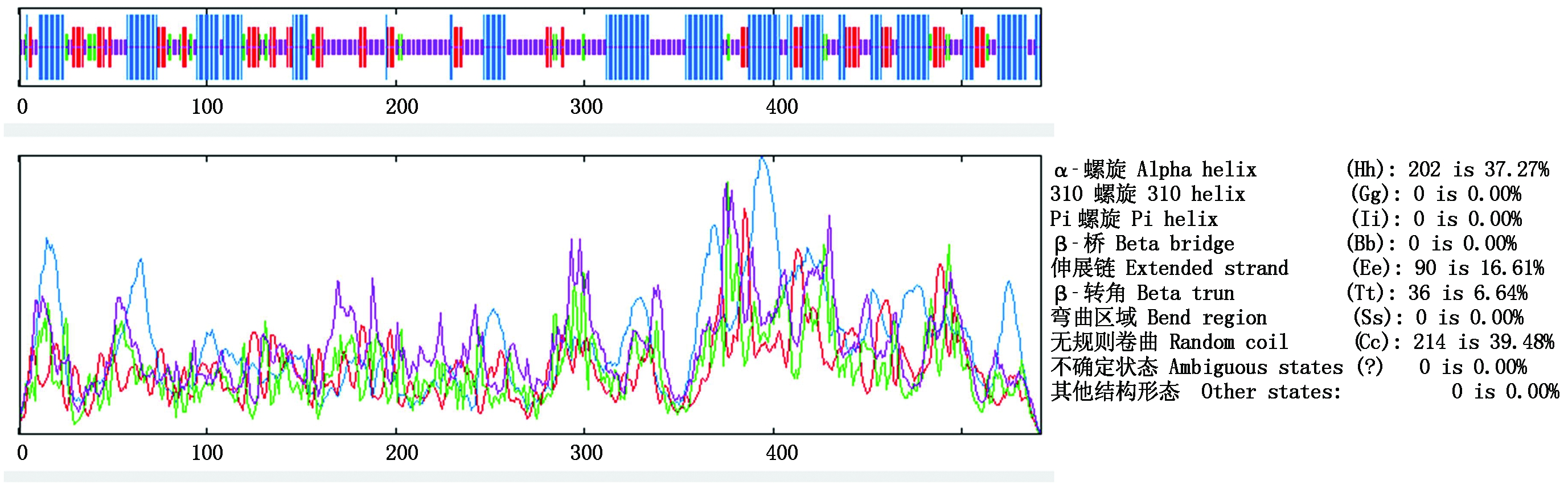
图1 GmbHLH041蛋白氨基酸序列的二级结构预测
Fig.1 The secondary structure prediction of the GmbHLH041 amino acids sequence
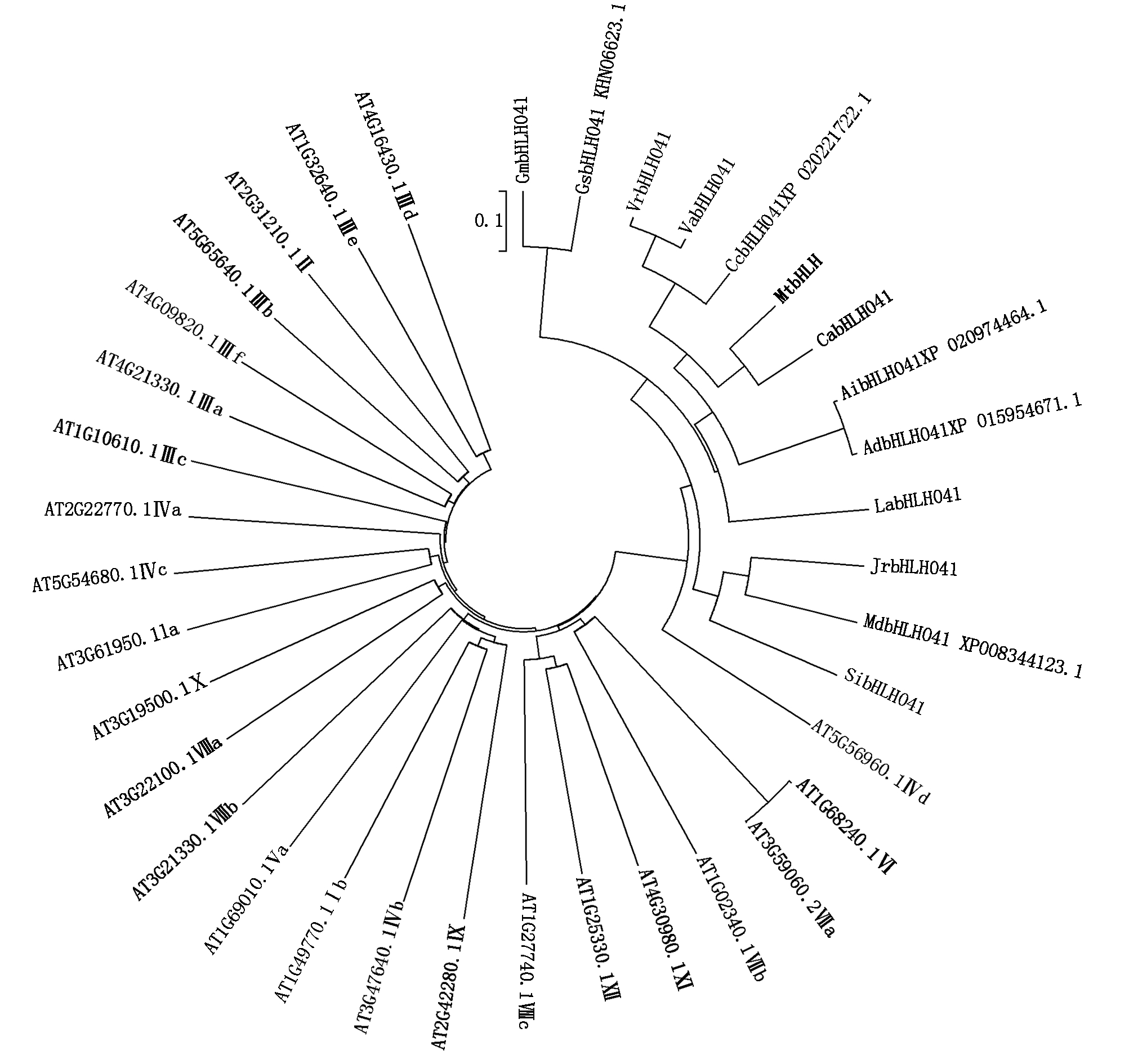
GmbHLH041. 大豆,XP_006600454.1;GsbHLH041.野生大豆,KHN06623.1;CcbHLH041.木豆,XP_020221722.1;VrbHLH041. 绿豆, XP_014500514.1;VabHLH041. 小豆, XP_017421300.1;LabHLH041. 狭叶羽扇豆,XP_019462343.1;MtbHLH. 蒺藜苜蓿, XP_003595200.2;CabHLH041. 鹰嘴豆, XP_004488061.1;AibHLH041. 花生,XP_020974464.1;AdbHLH041.蔓花生, XP_015954671.1;JrbHLH041.胡桃, XP_018845545.1;MdbHLH041.苹果, XP_008344123.1;SibHLH041. 芝麻, XP_011094007.1;AT5G56960.1 Ⅳd;AT1G68240.1Ⅴ1;AT3G59060.2Ⅶa;AT1G02340.1Ⅵb;AT4G30980.1X1;AT1G25330.1 Ⅻ;AT1G27740.1VⅢC;AT2G42280.1Ⅸ;AT3G47640.1Ⅴb;AT1G49770.1Ⅰb;AT1G69010.1 Ⅴa;AT3G21330.1Ⅷb;AT3G22100.1Ⅷa;AT3G19500.1Ⅹ;AT3G61950.1a;AT5G54680.1Ⅳc;AT2G22770.1Ⅳa;AT1G10610.1Ⅲc;AT4G21330.1Ⅲa;AT4G09820.1Ⅲf;AT5G65640.1Ⅲb;AT2G31210.1Ⅱ;AT1G32640.1Ⅲe;AT4G16430.1Ⅲd.拟南芥bHLH 12个亚家族中的24个bHLH蛋白。
GmbHLH041.(Glycine max,XP_006600454.1);GsbHLH041.(Glycine soja,KHN06623.1);CcbHLH041.(Cajanus cajan,XP_020221722.1);VrbHLH041.(Vigna radiata var. radiate, XP_014500514.1);VabHLH041.(Vigna angularis, XP_017421300.1);LabHLH041.(Lupinus angustifolius ,XP_019462343.1);MtbHLH.(Medicago truncatula, XP_003595200.2);CabHLH041.(Cicer arietinum, XP_004488061.1);AibHLH041.(Arachis ipaensis,XP_020974464.1);AdbHLH041.(Arachis duranensis, XP_015954671.1);JrbHLH041.(Juglans regia, XP_018845545.1);MdbHLH041.(Malus domestica, XP_008344123.1);SibHLH041.(Sesamum indicum, XP_011094007.1);AT5G56960.1 Ⅳd;AT1G68240.1Ⅴ1;AT3G59060.2Ⅶa;AT1G02340.1Ⅵb;AT4G30980.1Ⅹ1;AT1G25330.1 Ⅻ;AT1G27740.1ⅧC;AT2G42280.1Ⅸ;AT3G47640.1Vb;AT1G49770.1Ⅰb;AT1G69010.1 Ⅴa;AT3G21330.1Ⅷb;AT3G22100.1Ⅷa;AT3G19500.1Ⅹ;AT3G61950.1a;AT5G54680.1Ⅳc;AT2G22770.1Ⅳa;AT1G10610.1Ⅲc;AT4G21330.1Ⅲa;AT4G09820.1Ⅲf;AT5G65640.1Ⅲb;AT2G31210.1Ⅱ;AT1G32640.1Ⅲe;AT4G16430.1Ⅲd.24 Bhlh proteins from 12 bHLH subgroups of Arabidopsis thaliala.
图2 大豆GmbHLH041蛋白和其他物种bHLH蛋白的系统进化树分析
Fig.2 Phylogenetic tree analysis of soybean GmbHLH041 protein and other species bHLH proteins
2.2 大豆GmbHLH041基因编码蛋白系统进化树及保守结构序列分析
GmbHLH041蛋白在NCBI中进行比对,挑选亲缘关系较近的12个其他物种的bHLH,并根据拟南芥中bHLH转录因子亚家族的划分[16],在每个亚家族中选取至少 1个bHLH 氨基酸序列作为该亚家族的代表,从12个亚家族中共选取了24个 bHLH 蛋白质的氨基酸序列,进行进化树分析,结果显示,GmbHLH041与野生大豆(Glycine soja)和狭叶羽扇豆(Lupinus angustifolius)的bHLH041蛋白亲缘关系较近,属于拟南芥bHLH家族第4亚族Ⅳd中(图2)。
多重比对结果发现:GmbHLH041含有一个由60~70个氨基酸组成的保守的bHLH结构域,该结构域包括2个不同功能的保守区域:一个是位于N端参与DNA结合的由15个左右碱性氨基酸组成的区域(The basic region),另一个是位于C端的HLH区域(The helix-loop-helix region),主要参与形成二聚体,由2个疏水氨基酸形成的α-螺旋和1个可变环连接而成(图3)。
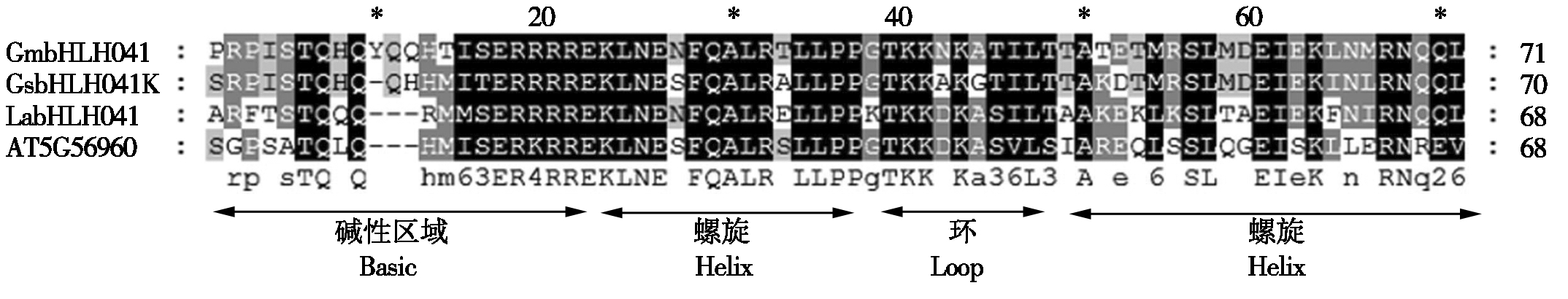
图3 大豆GmbHLH041蛋白保守序列多重比对
Fig.3 Comparison of amino acid sequences of the bHLH domains of GmbHLH041
2.3 GmbHLH041基因qRT-PCR分析
分析了大豆根系在50 μmol/L CdSO4胁迫24 h后GmbHLH041基因的表达情况,从图4可以看出,根系中GmbHLH041基因在Cd胁迫后表达量极显著上升。
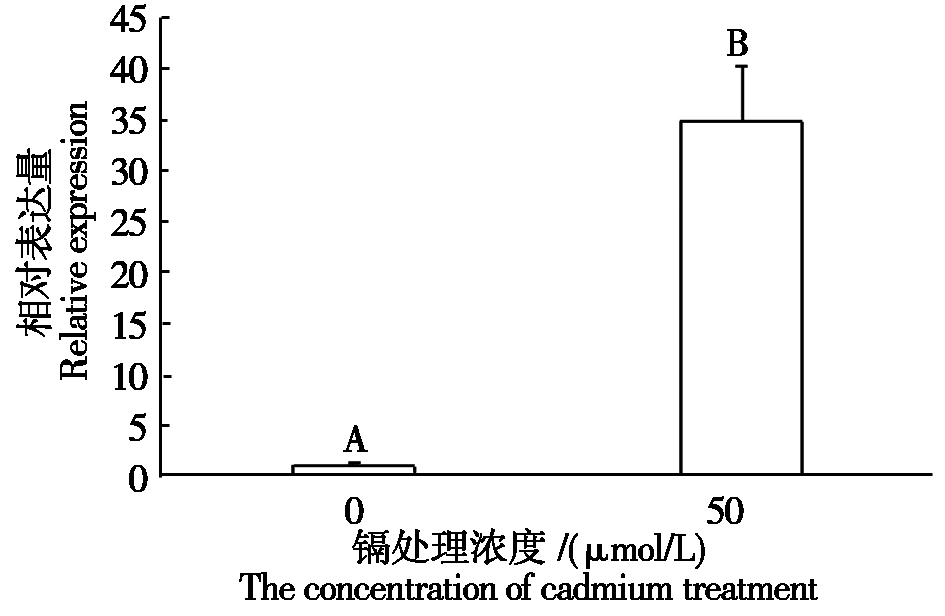
图中不同字母代表差异显著性(P<0.01)。
Different letters in the figure represent significant difference(P<0.01).
图4 50 μmol/L CdSO4胁迫24 h GmbHLH041基因在大豆根系中的表达情况
Fig.4 The expression of GmbHLH041 in soybean roots treated with 50 μmol/L CdSO4 for 24 h
3 讨论
镉是植物非必需营养元素,具有很强的生物毒性和较强的化学活性,在植物体内大量积累,会通过食物链危及人类的健康[2-3,17]。植物在进化过程中会形成一系列机制以应对环境胁迫[2]。
bHLH基因家族广泛参与植物的生长发育[18-19]、应对环境胁迫以及信号通路的调控[20-21]。目前,关于bHLH的研究主要集中在模式植物拟南芥中,近年来,陆续有研究报道,bHLH转录因子参与调控植物对金属离子的吸收。Van等[22]报道,Cd胁迫后,拟南芥以及Zn/Cd超富集植物天蓝遏蓝菜(Thlaspi caerulescens)中bHLH100转录因子的表达量上升,在拟南芥中过表达bHLH100能增加转基因植株对Zn和Ni的耐受性。最新的研究发现,3个拟南芥bHLH转录因子(FIT、AtbHLH38和AtbHLH39)参与了植物对Cd胁迫的响应[10]。
前期对Cd胁迫不同时间的大豆根系进行了转录组测序,对测序结果进行差异基因的筛选,发现GmbHLH041基因在Cd胁迫后表达量极显著上升。通过PCR从大豆根系中克隆到该基因,结合生物信息学预测蛋白结构,分析结果显示,该基因含有一个长度为1 629 bp的开放阅读框(Opening reading frame,ORF),编码542个氨基酸的蛋白质,GmbHLH041蛋白相对分子质量为61.17 ku,理论等电点为5.89,位于大豆第17条染色体上。与拟南芥的系统进化树分析显示,GmbHLH041属于第4亚族Ⅳd中,对4个bHLH同源基因氨基酸序列分析结果显示,GmbHLH041含有一个典型的bHLH结构域。荧光定量PCR结果表明,GmbHLH041基因在Cd胁迫后的大豆根系中上调表达,表明该基因可能与镉胁迫调控有关。已经进一步构建了表达载体,以期获得转基因植株,进一步研究该基因的生物学功能。
参考文献:
[1] Fang X,Wang L,Deng X,et al. Genome-wide characterization of soybean P1B-ATPasesgene family provides functional implications in cadmium responses[J]. BMC Genomics,2016,17(1):376.
[2] Sun N,Liu M,Zhang W T,et al. Bean Metal-responsive element-binding transcription factor confers cadmium resistance in tobacco[J]. Plant Physiology,2015,167(3):1136.
[3] Clemens S,Aarts M M,Thomine S,et al. Plant science:the key to preventing slow cadmium poisoning[J]. Trends in Plant Science,2013,18(2):92.
[4] Albert Q,Leleyter L,Lemoine M,et al. Determination and validation of soil thresholds for cadmium based on food quality standard and health risk assessment [J]. Science of the total environment,2018,619:700-706.
[5] Troise A D,Wiltafsky M,Fogliano V A. The quantification of free amadori compounds and amino acids allows to model the bound maillard reaction products formation in soybean products[J]. Food Chemistry,2018,247:29-38.
[6] Zatybekov A,Abugalieva S,Didorenko S A,et al. GWAS of agronomic traits in soybean collection included in breeding pool in Kazakhstan[J]. BMC Plant Biology,2017,17(1):179.
[7] Shute T,Macfie S M. Cadmium and zinc accumulation in soybean:a threat to food safety [J]. Science of the Total Environment,2006,371(1/3):63-73.
[8] Hall J L,Williams L E. Transition metal transporters in plants[J]. Journal of Experimental Botany,2003,54(393):2601-2613.
[9] Krämer U,Talke I N,Hanikenne M. Transition metal transport[J]. FEBS Letters,2007,581(12):2263-2272.
[10] Wu H,Chen C,Du J,et al. Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots[J]. Plant Physiology,2012,158(2):790.
[11] Ledent V,Vervoort M. The basic helix-loop-helix protein family:comparative genomics and phylogenetic analysis[J]. Genome Research,2001,11(5):754.
[12] Li X L,Zhang H M,Ai Q,et al. Two bHLH transcription factors,bHLH34 and bHLH104,regulate Iron homeostasis in Arabidopsis thaliana[J]. Plant Physiology,2016,170(4):2478-2493.
[13] Amoutzias G D,Robertson D L,Oliver S G,et al. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes[J]. EMBO Reports,2004,5(3):274-279.
[14] Castillon A,Shen H,Huq E. Phytochrome interacting factors:central players in phytochrome-mediated light signaling networks[J]. Trends in Plant Science,2007,12(11):514-521.
[15] Stevens J D,Roalson E H,Skinner M K. Phylogenetic and expression analysis of the basic helix-loop-helix transcription factor gene family:genomic approach to cellular differentiation[J]. Differentiation,2008,76(9):1006-1022.
[16] Heim M A,Jakoby M,Werber M,et al. The basic helix-loop-helix transcription factor family in plants:a genome-wide study of protein structure and functional diversity[J]. Molecular Biology and Evolution,2003,20(5):735-747.
[17] Fang X L,Wang L,Deng X J,et al. Genome-wide characterization of soybean P-1B-ATPase gene family provides functional implications in cadmium responses[J]. BMC Genomics,2016,17:376.
[18] Zhu Z G,Chen G P,Guo X H,et al. Overexpression of SlPRE2,an atypical bHLH transcription factor,affects plant morphology and fruit pigment accumulation in tomato[J]. Scientific Reports,2017,7(1):5786.
[19] Moerkercke A V,Steensma P,Schweizer F,et al. The bHLH transcription factor BIS1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus[J]. Proceedings of the National Academy of Science,2015,112(26):8130-8135.
[20] Patra B,Pattanaik S,Schluttenhofer C,et al. A network of jasmonate-responsive bHLH factors modulate monoterpenoid indole alkaloid biosynthesis in Catharanthus roseus[J]. New Phytologist,2017,217(4):1566-1581.
[21] Takahashi Y,Ebisu Y,Shimazaki K I. Reconstitution of abscisic acid signaling from the receptor to DNA via bHLH transcription factors[J]. Plant Physiology,2017,174(2):815.
[22] Van D M,Judith E,Schat H,et al. Expression differences for genes involved in lignin,glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens[J]. Plant,Cell & Environment,2008,31(3):301-324.