灰霉病是一种世界性分布的植物病害,其病原为灰葡萄孢(Botrytis cinerea),主要为害幼果以及成熟的果实、花序、叶片、果柄等,常导致花序、果实大量脱落,已成为严重影响我国大田和温室作物产量和品质的重大病害。目前,灰霉病的防治仍以化学防治为主,但由于灰葡萄孢具有遗传变异大、繁殖速度快、适应性强以及具有多次再侵染等特点,使其对多种不同作用机制的杀菌剂产生了抗药性,导致杀菌剂的使用量逐年增大,并对农产品的安全性造成了严重威胁。因此,挖掘植物抗灰霉病基因,深入研究其抗病分子机制,为培育抗灰霉病的作物新品种具有重要意义。近年来,从拟南芥中克隆得到的抗病基因以及抗性相关基因一直是人们研究的热点。目前,大部分植物抗病基因及抗性相关基因是从拟南芥中获得的,如RPS2[1-2]、RPS5、RPS6[3]、RPM1[1,4-5]、RPP4[6-7]、RPP7[8]、RPW8[9-12]等,其中部分抗病基因已经应用到生产上。研究显示,植物产生抵抗死体病原菌灰葡萄孢的侵染时,水杨酸(SA)信号途径发挥了重要作用,SA的积累可以增加植物对灰葡萄孢的局部抗性[13]。通过高通量转录组分析,已经鉴别了大量植物抵抗灰葡萄孢侵染起作用的转录因子TFs(Transcription factors)[14-17]。拟南芥T1N6_22基因编码的蛋白为NAD(P)结合Rossmann-折叠蛋白家族的成员,具有葡萄糖脱氢酶活性和短链氧化还原酶活性,参与植物体内各种催化反应和新陈代谢过程,已经确定其通过参与SA和茉莉酸(JA)信号途径调控拟南芥对灰葡萄孢的抗性[18]。河北农业大学真菌毒素与植物分子病理学实验室前期获得了一株对灰葡萄孢敏感的拟南芥突变体,确定了其突变基因为T1N6_22;通过互补回复试验,明确了T1N6_22基因在拟南芥抗灰霉病过程中起正调控作用,在拟南芥抗丁香假单胞杆菌(Pst DC3000)过程中起负调控作用;利用酵母双杂交技术,以T1N6_22蛋白为诱饵筛选拟南芥的酵母cDNA文库,获得了T1N6_22蛋白的候选互作蛋白[19]。但是,T1N6_22蛋白的互作蛋白及其调控拟南芥抗病的分子机制尚未明确。
本研究利用酵母双杂交技术,对拟南芥抗病相关基因T1N6_22的互作蛋白进行鉴定,旨在为进一步明确T1N6_22基因调控拟南芥抗病的分子机制奠定基础。
1 材料和方法
1.1 供试材料
酵母双杂交载体PGADT7(AD)和PGBKT7(BD)、酵母菌株AH109、拟南芥Columbia生态型(Col-0)均由河北省植物生理与分子病理学重点实验室、河北农业大学真菌毒素与植物分子病理学实验室提供;酵母转化试剂和酵母培养基购于美国Clontech公司;pCR8克隆试剂盒(K2520-20)和LR克隆试剂盒(11791020)购于美国Invitrogen公司。
1.2 T1N6_22基因及其候选互作蛋白基因的扩增
提取拟南芥Columbia生态型(Col-0)植株的总RNA,总RNA提取方法参照提取试剂盒OMEGA Plant RNA Kit的说明书。以提取的总RNA为模板,反转录合成cDNA,cDNA的合成方法参照宝生物反转录试剂盒的说明书。以cDNA为模板,使用T1N6_22基因和AT1G06050、AT1G21400、AT2G19480基因的特异引物进行PCR扩增(表1)。PCR反应体系为MgSO4 2 μL、10×PCR Buffer 5 μL、dNTP(2.5 mmol/L)1 μL、引物各1 μL、模板cDNA 2 μL、Taq酶0.2 μL,最后加ddH2O至50 μL。PCR程序为94 ℃ 2 min;94 ℃ 15 s,56 ℃ 30 s,72 ℃ 1 min,35个循环。用1%琼脂糖凝胶电泳检测PCR产物。
表1 引物序列
Tab.1 Primer sequence

基因名称Genename引物序列(5′-3′)PrimersequenceT1N6_22ATGGCAGACCCAAGAGTTGCAGTTCAAAAGTTGGAGACATTGGCGCAT1G06050ATGGCTGGCTCTGTTGGTGTTACTGCTTCTGTACATGTTCATCCAT1G21400ATGGCGATCTGGTTTGCTAGTCAAACATGAAAGCCAGGAGAT2G19480ATGAGCAACGACAAGGACAGCTCACTGCTGCTTACATTCCGG
1.3 T1N6_22基因及其候选互作蛋白基因的克隆
将T1N6_22基因及其候选互作蛋白基因的PCR扩增产物进行回收,胶回收方法参照全式金胶回收试剂盒的说明书。将回收的基因扩增产物分别与Gateway克隆载体pCR8进行连接。连接体系为1 μL回收的PCR产物、盐溶液0.5 μL、ddH2O 1 μL、pCR8 Topo载体0.5 μL,室温23 ℃反应5 min。连接后转化大肠杆菌DH5α,挑取阳性克隆进行PCR鉴定和测序验证。
1.4 T1N6_22基因及其候选互作蛋白基因的酵母双杂交载体构建
利用质粒提取试剂盒,提取经测序正确的pCR8-T1N6_22、pCR8-AT1G06050、pCR8-AT1G21400和pCR8-AT2G19480质粒;利用LR重组试剂盒,将4个基因的入门载体分别与酵母双杂交载体PGADT7(AD)和PGBKT7(BD)进行重组反应,反应体系为质粒pCR8-T1N6_22(或者pCR8-AT1G06050、pCR8-AT1G21400、pCR8-AT2G19480)3 μL、AD/BD载体1 μL、LR克隆酶Ⅱ 1 μL,25 ℃连接2 h。2 h后在连接体系中加入蛋白酶K 1 μL,37 ℃ 10 min。将所得到的连接产物转化大肠杆菌DH5α,挑取阳性克隆进行PCR检测,将PCR检测正确的克隆进行进一步的测序鉴定。
1.5 T1N6_22基因及其候选互作蛋白基因的自激活活性检测
将1 μg AD质粒分别与1 μg BD-T1N6_22、BD-AT1G06050、BD-AT1G21400和BD-AT2G19480组合共同转化酵母感受态细胞AH109,室温孵育1~2 h,42 ℃热激30 min后,冰浴1~2 min。将酵母细胞涂布于二缺(-Leu/-Trp)培养基上,30 ℃培养2~3 d。挑取二缺培养基上生长良好的酵母单克隆,用100 μL无菌水稀释,吸取10 μL点于三缺-Leu/-Trp/-His和添加3-AT的-Leu/-Trp/-His培养基上,30 ℃继续培养2~3 d,观察酵母生长情况。
1.6 酵母双杂交
将1 μg AD-T1N6_22质粒分别与1 μg BD-AT1G06050、BD-AT1G21400和BD-AT2G19480质粒组合,将1 μg BD-T1N6_22质粒分别与1 μg AD-AT1G06050、AD-AT1G21400和AD-AT2G19480质粒组合,同时设对照组合AD-T1N6_22+BD、BD-AT1G06050+AD、BD-AT1G21400+AD、BD-AT2G19480+AD、BD-T1N6_22+AD、AD-AT1G06050+BD、AD-AT1G21400+BD和AD-AT2G19480+BD。将不同组合分别转化酵母感受态细胞(200 μL),涂布二缺(-Leu/-Trp)培养基上,30 ℃培养2~3 d。挑取二缺培养基上生长良好的酵母单克隆,用100 μL无菌水稀释,分别吸取10 μL点于二缺-Leu/-Trp、三缺-Leu/-Trp/-His、添加3-AT的三缺-Leu/-Trp/-His 3-AT和四缺-Leu/-Trp/-His/-Ade培养基上,30 ℃继续培养2~3 d,观察酵母生长情况。
2 结果与分析
2.1 T1N6_22基因及其候选互作蛋白基因的酵母双杂交载体构建
利用T1N6_22基因的特异性引物扩增得到T1N6_22基因的全长(888 bp)(图1-A),将其与入门载体pCR8连接后转化大肠杆菌DH5α,经菌落PCR扩增检测发现获得了单一的目的条带(图1-B),进一步对其进行测序验证,获得T1N6_22基因入门载体pCR8-T1N6_22。将pCR8-T1N6_22与酵母双杂交载体AD、BD进行重组反应,反应产物转化大肠杆菌DH5α,经菌落PCR鉴定获得了单一的目的条带(图1-C、D),进一步对重组载体进行测序鉴定,最终获得T1N6_22基因酵母双杂交载体AD-T1N6_22和BD-T1N6_22。利用同样的方法,分别获得AT1G06050、AT1G21400和AT2G19480基因的酵母双杂交载体AD-AT1G06050、BD-AT1G06050、AD-AT1G2140、BD-AT1G2140、AD-AT2G19480、BD-AT2G19480(图2-4)。
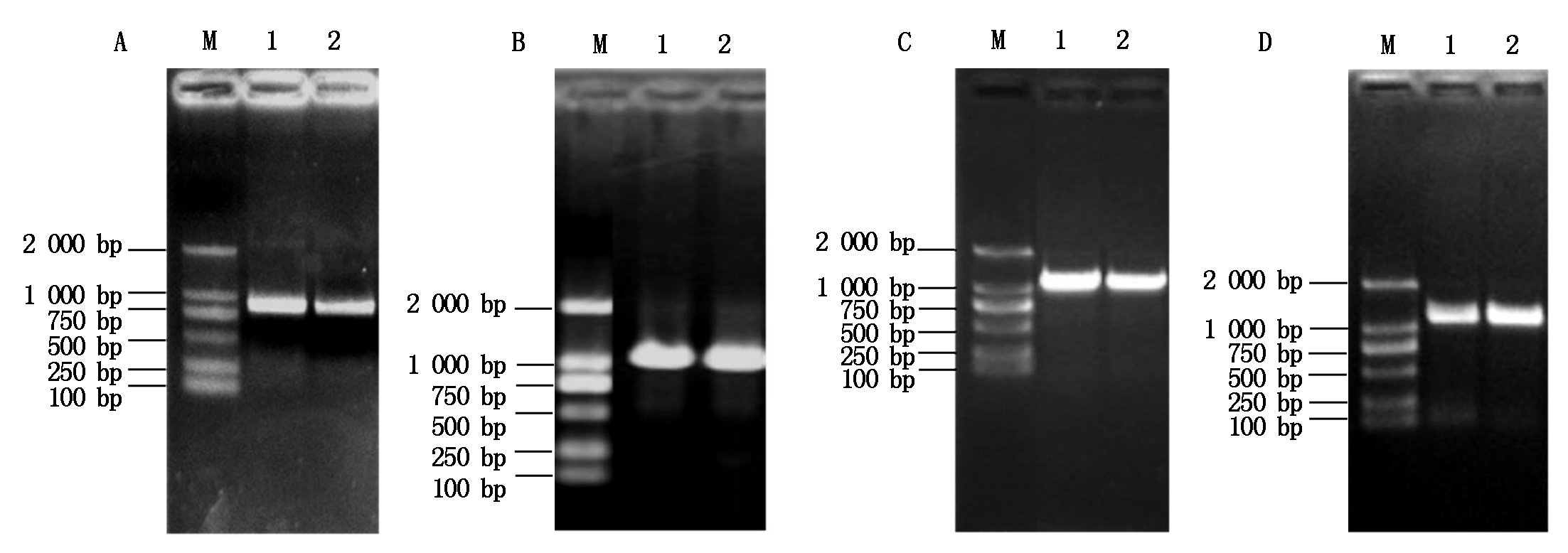
A.T1N6_22的PCR扩增;B.pCR8-T1N6_22的PCR扩增;C.AD-T1N6_22的PCR扩增;D.BD-T1N6_22的PCR扩增。A.PCR amplification of T1N6_22;B.PCR amplification of pCR8-T1N6_22;C.PCR amplification of AD-T1N6_22;D.PCR amplification of BD-T1N6_22.
图1 T1N6_22基因酵母双杂交载体的构建
Fig.1 Construction of the T1N6_22 gene yeast two-hybrid vector
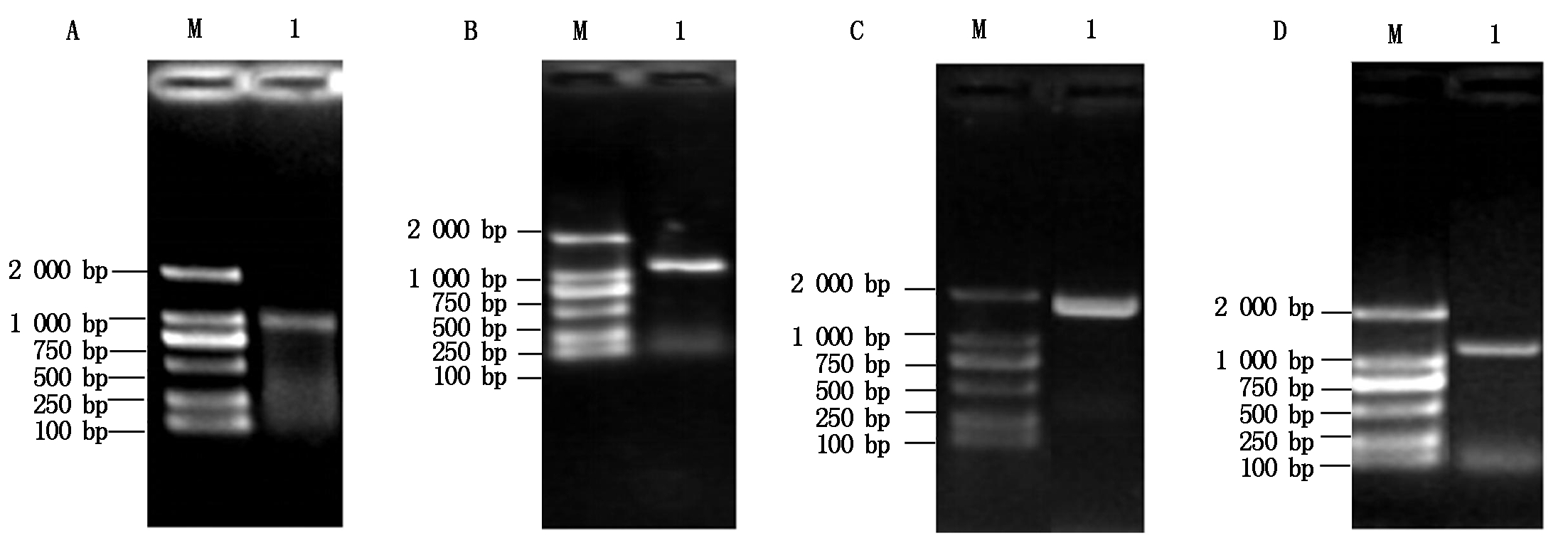
A.AT1G06050的PCR扩增;B.pCR8-AT1G06050的PCR扩增;C.AD-AT1G06050的PCR扩增;D.BD-AT1G06050的PCR扩增。A.PCR amplification of AT1G06050;B.PCR amplification of pCR8-AT1G06050;C. PCR amplification of the AD-AT1G06050;D. PCR amplification of BD-AT1G06050.
图2 AT1G06050基因酵母双杂交载体的构建
Fig.2 Construction of the AT1G06050 gene yeast two-hybrid vector
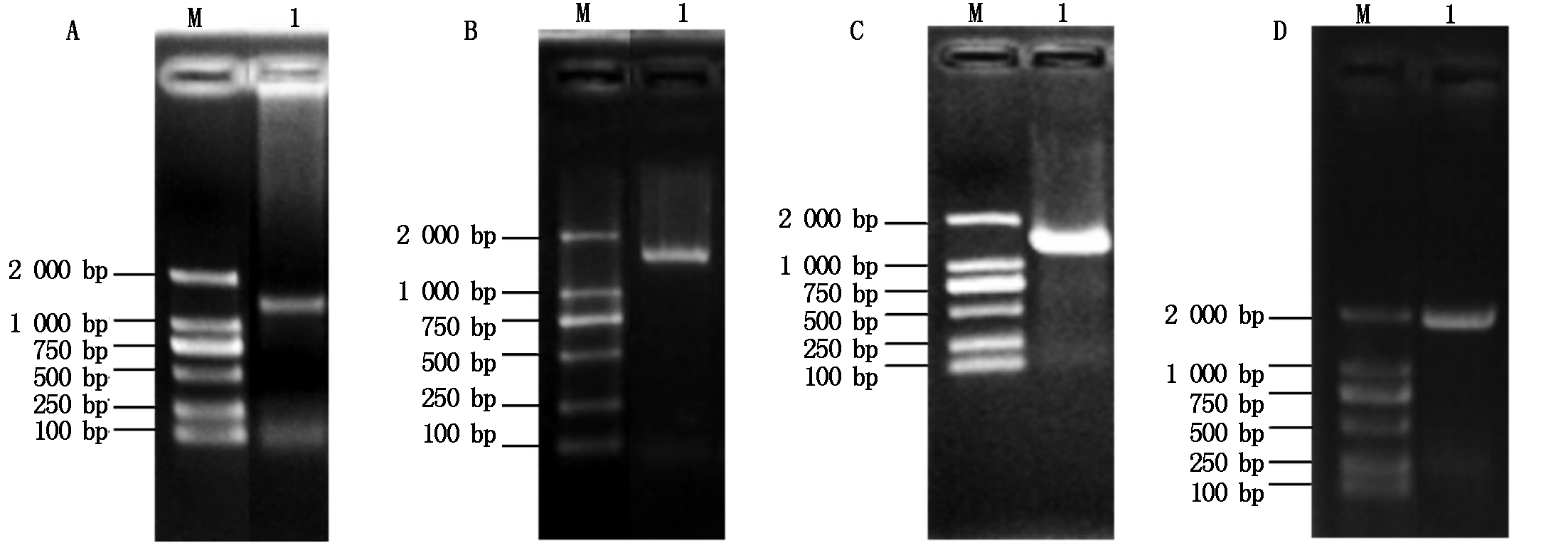
A.AT1G21400的PCR扩增;B.pCR8-AT1G21400的PCR扩增;C.AD-AT1G21400的PCR扩增;D.BD-AT1G21400的PCR扩增。A.PCR amplification of AT1G21400;B.PCR amplification of pCR8-AT1G21400;C.PCR amplification of AD-AT1G21400;D.PCR amplification of BD-AT1G21400.
图3 AT1G21400基因酵母双杂交载体的构建
Fig.3 Construction of the AT1G21400 gene yeast two-hybrid vector
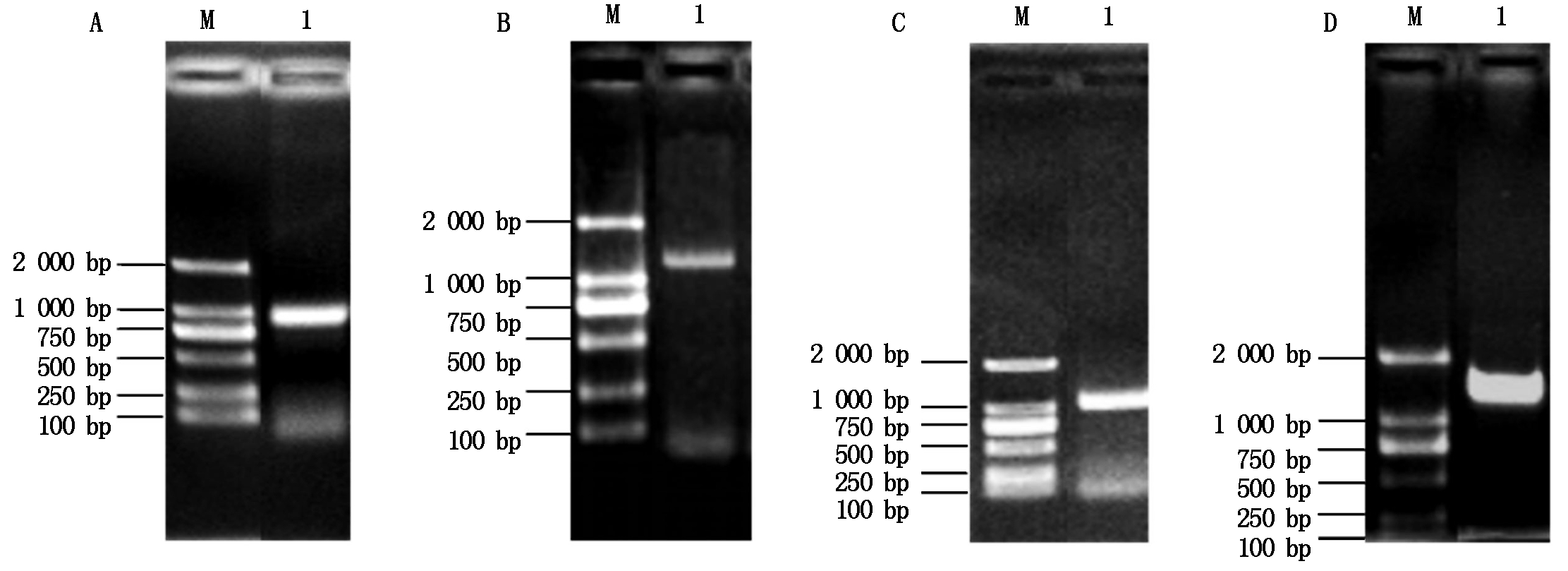
A.AT2G19480的PCR扩增;B.pCR8-AT2G19480的PCR扩增;C.AD-AT2G19480的PCR扩增;D.BD-AT2G19480的PCR扩增。A.PCR amplification of AT2G19480;B. PCR amplification of pCR8-AT2G19480;C. PCR amplification of AD-AT2G19480;D. PCR amplification of BD-AT2G19480.
图4 AT2G19480基因酵母双杂交载体的构建
Fig.4 Construction of AT2G19480 gene yeast two-hybrid vector
2.2 T1N6_22基因及其候选互作蛋白基因的自激活活性检测
将AD空载体分别与BD-T1N6_22、BD-AT1G06050、BD-AT1G21400和BD-AT2G19480质粒组合共转化酵母感受态细胞,检测各基因的自激活活性。结果发现,共同转化AD与BD-T1N6_22、BD-AT1G21400和BD-AT2G19480载体的酵母菌落均不能在三缺培养基-Leu/-Trp/-His和-Leu/-Trp/-His 3-AT上生长,共同转化BD-AT1G06050和AD载体的酵母菌落能在三缺(-Leu/-Trp/-His)培养基上正常生长,添加3-AT的三缺培养基-Leu/-Trp/-His 3-AT能够抑制其生长(图5)。表明T1N6_22、AT1G21400、AT2G19480基因无自激活活性,AT1G06050基因有自激活活性。
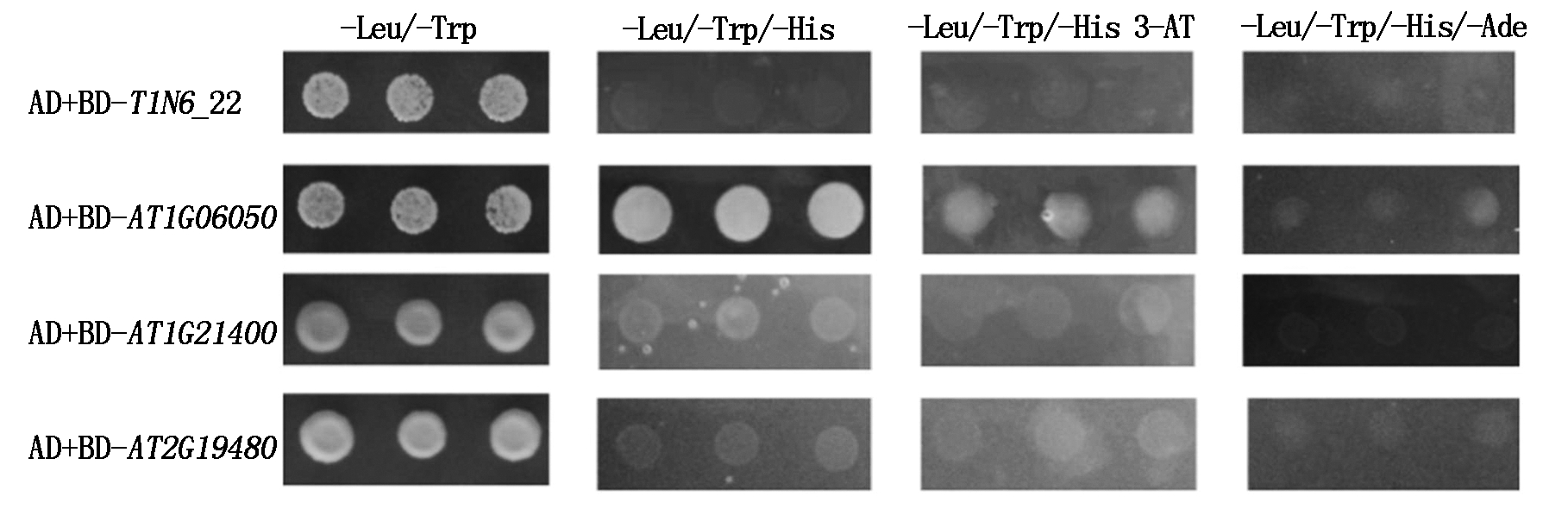
图 5 T1N6_22及其可能互作蛋白自激活活性的鉴定
Fig.5 Self-activating activity of T1N6_22 and its candidate interacting protein
2.3 T1N6_22互作蛋白的酵母双杂交鉴定
将AD-T1N6_22与BD-AT1G06050、BD-T1N6_22与AD-AT1G06050组合进行酵母双杂交试验,同时设AD-T1N6_22+BD、AD+BD-AT1G06050、BD-T1N6_22+AD、BD+AD-AT1G06050、AD+BD组合作为阴性对照。结果发现,共转化AD-T1N6_22与BD-AT1G06050、BD-T1N6_22与AD-AT1G06050组合的酵母菌落在三缺培养基(-Leu/-Trp/-His和-Leu/-Trp/-His 3-AT)和四缺培养基(-Leu/-Trp/-His/-Ade)均能生长,共转化阴性对照AD与BD-AT1G06050组合的酵母菌落在三缺培养基(-Leu/-Trp/-His)上能够生长,而共转化其他阴性对照组合的酵母菌落在三缺培养基(-Leu/-Trp/-His和-Leu/-Trp/-His 3-AT)和四缺培养基(-Leu/-Trp/-His/-Ade)上均不能生长(图6)。表明T1N6_22与AT1G06050能在酵母细胞中直接互作。
利用同样的方法,检测T1N6_22与AT1G21400、AT2G19480之间在酵母中的互作关系,结果发现,共转化AD-T1N6_22+BD-AT1G21400、BD-T1N6_22+AD-AT1G21400、AD-T1N6_22+BD-AT2G19480、BD-T1N6_22+AD-AT2G19480组合的酵母菌落在三缺培养基(-Leu/-Trp/-His和-Leu/-Trp/-His 3-AT)和四缺培养基(-Leu/-Trp/-His/-Ade)上均能生长,而阴性对照组合除BD+AD-AT1G21400和AD+BD在三缺培养基(-Leu/-Trp/-His)上有微弱生长外,其余阴性对照组合均不能在三缺和四缺培养基上正常生长(图7-8)。表明T1N6_22与AT1G21400、AT2G19480能在酵母细胞中直接互作。
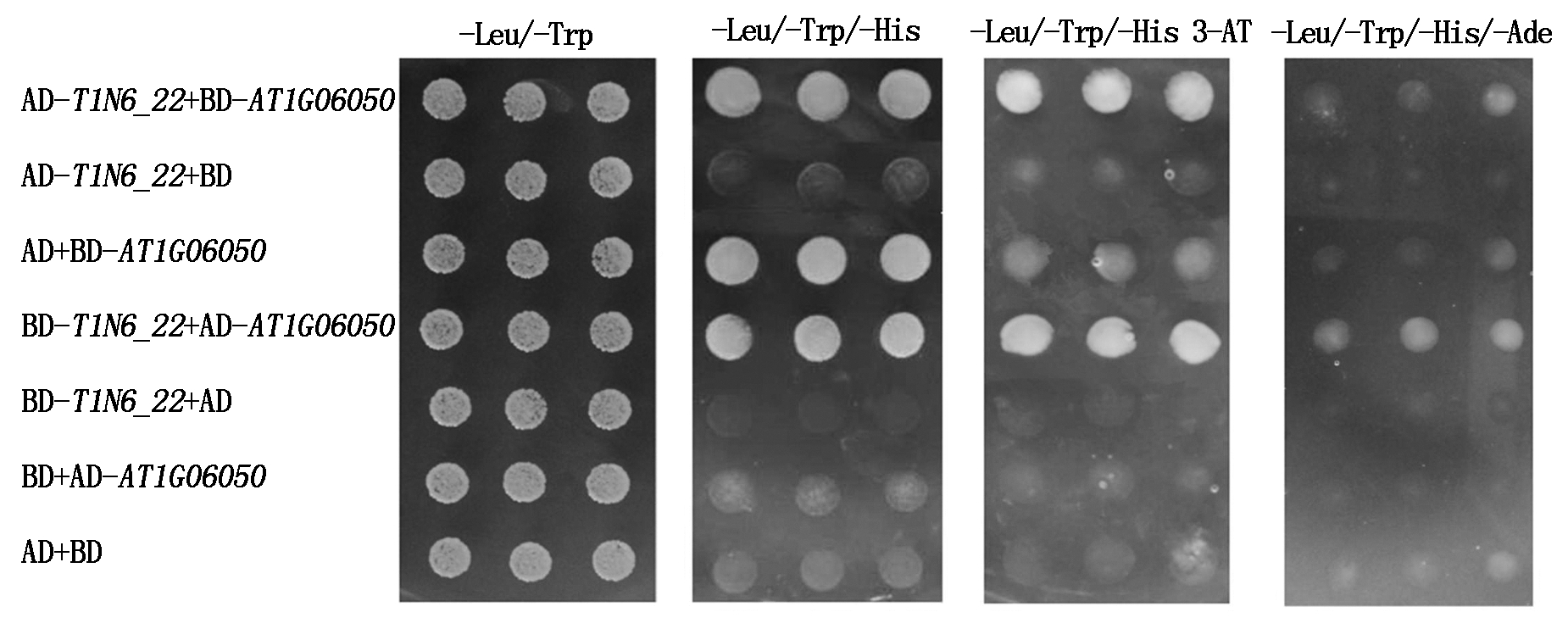
图 6 T1N6_22 和AT1G06050酵母双杂交
Fig.6 Results of T1N6_22 and AT1G06050 yeast two-hybrid
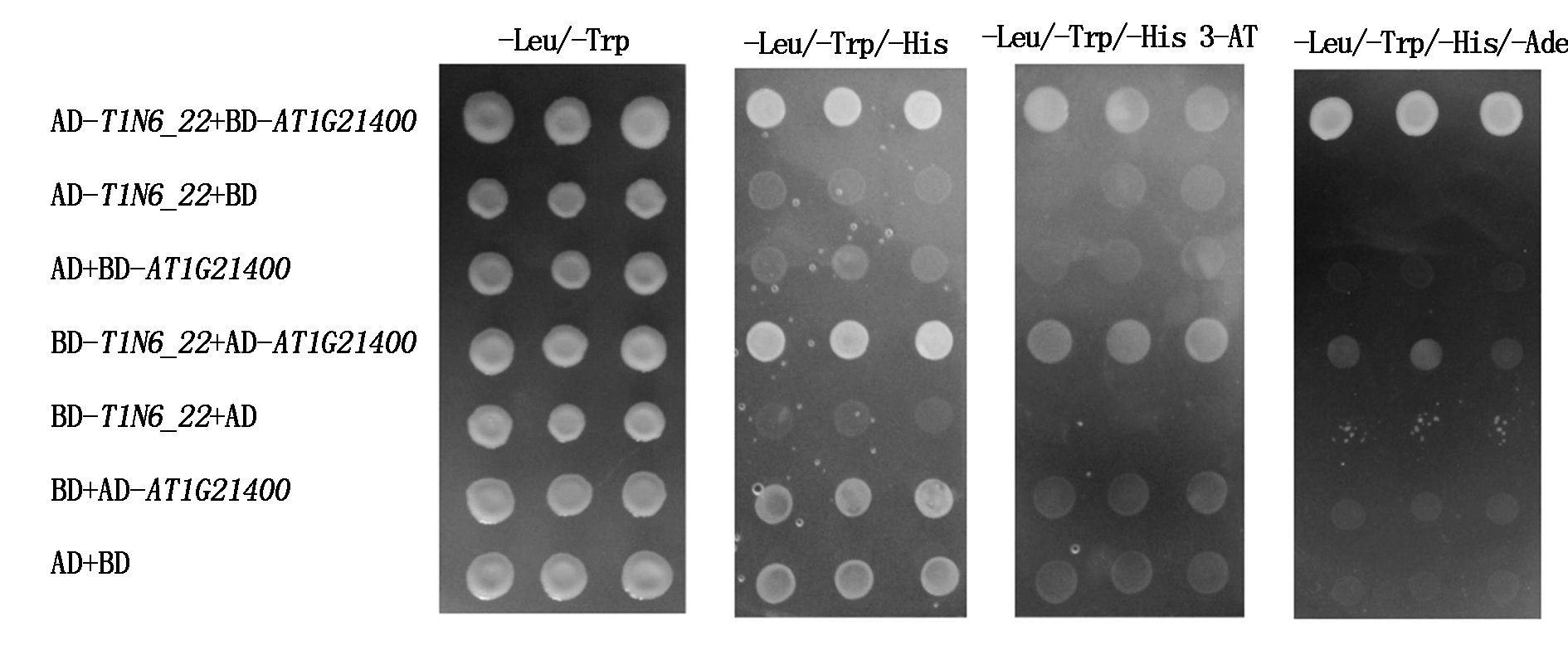
图7 T1N6_22 和AT1G21400酵母双杂交
Fig.7 Results of T1N6_22 and AT1G21400 yeast two-hybrid
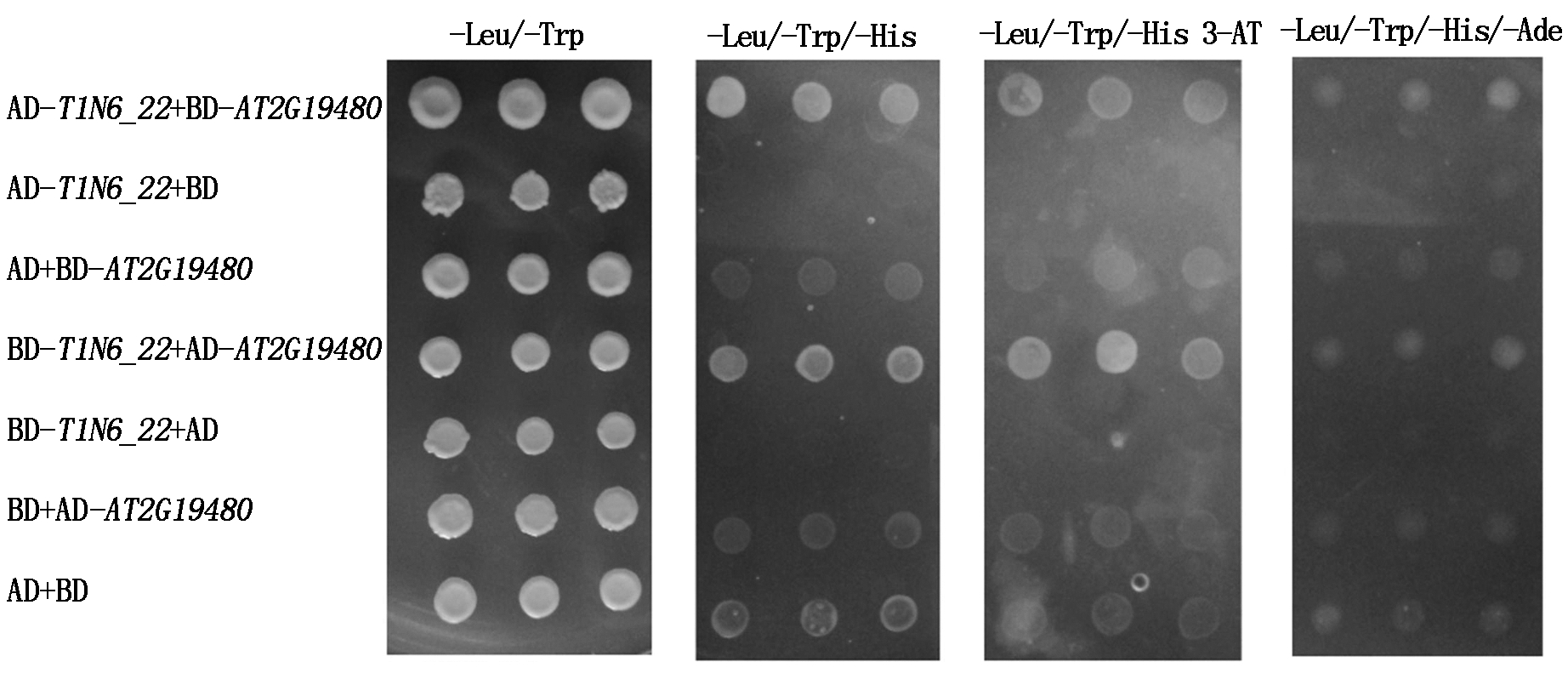
图8 T1N6_22 和AT2G19480酵母双杂交
Fig.8 Results of T1N6_22 and AT2G19480 yeast two-hybrid
3 讨论
酵母双杂交系统(Yeast two-hybrid system)是分析蛋白与蛋白之间相互作用的有效、快速的方法,可以精确地测定蛋白质之间微弱的相互作用,由于其操作水平是在核酸水平,不需要纯化大量的蛋白,因此,操作简单容易。但是,酵母双杂交技术存在一些自身缺陷,很明显的一个缺陷就是存在假阳性。因此,酵母双杂交验证的蛋白质相互作用往往还需要其他的试验证据进一步支持。本研究中,利用酵母双杂交技术确定了T1N6_22蛋白与AT1G06050、AT1G21400和AT2G19480存在互作关系。基于酵母双杂交系统自身的局限性,后续试验中可以采用其他技术对其互作关系进行进一步的验证。
除了酵母双杂交之外,还有多种检测蛋白互作的方法。如双分子荧光互补技术(BiFC)和免疫共沉淀技术(Co-IP)。其中,BiFC技术是一个在活细胞中检测蛋白互作的非常好的工具[20-23];Co-IP技术是一种在细胞非变性条件下研究蛋白之间直接互作的方法,可以在体内直接确定它们之间相互作用方式的动态变化[24]。
本研究利用酵母双杂交技术确定了T1N6_22蛋白的互作蛋白AT1G06050、AT1G21400和AT2G19480,下一步工作可以对AT1G06050、AT1G21400和AT2G19480的功能进行深入研究,明确其在拟南芥抗病中的功能及其与SA和JA信号途径之间的关系,深入探讨T1N6_22基因及其互作蛋白基因调控拟南芥抗病的分子机制奠定基础。
参考文献:
[1] Belkhadir Y,Nimchuk Z,Hubert D A,et al. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type Ⅲ effectors AvrRpt2 or AvrRpm1[J]. Plant Cell,2004,16(10):2822-2835.
[2] Takahashi A,Casais C,Ichimura K,et al. HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis[J]. Proceedings of the National Academy of Sciences of the United States of America,2003,100(20):11777-11782.
[3] Kim S H,Kwon S,Saha D,et al. Resistance to the pseudomonas syringae effector hopA1 is governed by the TIR-NBS-LRR protein RPS6 and is enhanced by mutations in SRFR1[J]. Plant Physiology,2009,150(4):1723-1732.
[4] Tornero P,Chao R,Luthin W,et al. Large-scale structure-function analysis of the Arabidopsis RPM1 disease resistance protein[J]. Plant Cell,2002,14(2):435-450.
[5] Serrano M,Hubert D,Dangl J L,et al. A chemical screen for suppressors of the avr Rpm1-RPM1-dependent hypersensitive cell death response in Arabidopsis thaliana[J]. Planta,2010,231(5):1013-1023.
[6] Xia S T,Cheng Y T,Huang S,et al. Regulation of transcription of Nucleotide-Binding Leucine-Rich Repeat-Encoding genes SNC1 and RPP4 via H3K4 trimethylation[J]. Plant Physiology,2013,162(3):1694-1705.
[7] Bao F,Huang X Z,Zhu C P,et al. Arabidopsis HSP90 protein modulates RPP4-mediated temperature-dependent cell death and defense responses[J]. New Phytologist,2014,202(4):1320-1334.
[8] Eulgem T,Tsuchiya T,Wang X J,et al. EDM2 is required for RPP7-dependent disease resistance in Arabidopsis and affects RPP7 transcript levels[J]. Plant Journal,2007,49(5):829-839.
[9] Wang W M,Ma X F,Zhang Y,et al. PAPP2C interacts with the atypical disease resistance protein RPW8.2 and negatively regulates salicylic Acid-Dependent defense responses in Arabidopsis[J]. Molecular Plant,2012,5(5):1125-1137.
[10] Wang W,Zhang Y,Wen Y,et al. A comprehensive mutational analysis of the Arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting[J]. Plant Cell,2013,25(10):4242-4261.
[11] Wang W,Berkey R,Wen Y,et al. Accurate and adequate spatiotemporal expression and localization of RPW8.2 is key to activation of resistance at the host-pathogen interface[J]. Plant Signaling & Behavior,2010,5(8):1002-1005.
[12] Kim H,O′connell R,Maekawa-Yoshikawa M A,et al. The powdery mildew resistance protein RPW8.2 is carried on VAMP721/722 vesicles to the extrahaustorial membrane of haustorial complexes[J]. Plant Journal,2014,79(5):835-847.
[13] Si-ammour A,Mauch-Mani B,Mauch F.Quantification of induced resistance against Phytophthora species expressing GFP as a vital marker:β-aminobutyric acid but not BTH protectspotato and Arabidopsis from infection[J]. Molecular Plant Pathology,2003,4(4):237-248.
[14] Ferrari S,Galletti R,Denoux C,et al. Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is idependent of salicylic acid,ethylene,or jasmonate signaling but requires PHYTOALEXIN DEFICIENT3[J]. Plant Physiology,2007,144(1):367-379.
[15] Rowe H,Walley J,Corwin J,et al. Deficiencies in jasmonate-mediated plant defensereveal quantitative variation in Botrytis cinerea pathogenesis[J]. PLoS Pathogens,2010,6(4):e1000861.
[16] Birkenbihl R,Diezel C,Somssich I. Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection[J]. Plant Physiology,2012,159(1):266-285.
[17] Mulema J,Denby K. Spatial and temporal transcriptomic analysis of the Arabidopsis thaliana-Botrytis cinerea interaction[J]. Molecular Biology Reports,2012,39(4):4039-4049.
[18] 邢继红,班卫红,瓮巧云,等. 拟南芥不同生态型对灰葡萄孢的抗性[J]. 河北农业大学学报,2010,6(6):26-30.
[19] 瓮巧云,黄聪聪,王 娜,等.拟南芥抗灰霉病基因T1N6_22互作蛋白的筛选与分析[J]. 华北农学报,2017,32(2):15-20.
[20] Nishimura K,Ishikawa S,Matsunami E,et al. New gateway-compatible vectors for a high-throughput protein-protein interaction analysis by a bimolecular fluorescence complementation (BiFC) assay in plants and their application to a plant clathrin structure analysis[J]. Bioscience Biotechnology and Biochemistry,2015,79(12):1995-2006.
[21] Wang J,Yu Y W,Zhang Z J,et al. Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis[J]. Plant Cell,2013,25(2):625-636.
[22] Song J W,Xing Y L,Munir S,et al. An ATL78-Like RING-H2 finger protein confers abiotic stress tolerance through interacting with RAV2 and CSN5B in tomato[J]. Frontiers in Plant Science,2016,7:1305.
[23] Thi T L C,Brumbarova T,Ivanov R,et al. Zinc finger of Arabidopsis thaliana 12 (zat12) interacts with fer-like iron deficiency-induced transcription factor (fit) linking iron deficiency and oxidative stress responses[J]. Plant Physiology,2016,170(1):540-557.
[24] 李 玲,杨鹏跃,朱本忠,等. 染色质免疫共沉淀技术的应用和研究进展[J]. 中国食品学报,2012,12(6):124-132.