盐胁迫是影响农作物生长最重要的非生物胁迫[1],随着全球变暖的到来,盐渍土的问题日趋严重,近些年来,由于化学肥料的施用和不合理的灌溉措施,更是快速加大了土壤盐渍化的进度[2],水稻是全世界重要的粮食作物,但由于栽培措施不合理导致的土壤盐渍化已经严重制约了水稻生产[3],在全球,大约30%的水稻种植区面临盐胁迫[4],且水稻的不同生长时期均有盐胁迫的发生,然而水稻耐盐机制尚未明确,因此很难开展耐盐水稻品种的研究[5]。
QTL分析是探索性状遗传机理的重要环节,在水稻耐盐QTL研究中,前人针对水稻不同生长时期做了大量报道[2,5-10],且基于初定位的QTL由于群体类型和大小存在区别,产生了不同的遗传效应,这些初步的研究很难将QTL与候选基因关联在一起,因为多数QTL的遗传距离很大,置信区间里包含了数以千计的基因[11],因此需要通过比较不同QTL,筛选出置信区间小且遗传效应高的一致性QTL。元分析能够把来自不同研究的QTL信息联系到一起,通过整合QTL信息和建模,达到优化QTL置信区间的目的,从而揭示不同性状间的遗传关系[12]。因此,近年来,棉花[13]、黑麦草[14]、玉米[15]、花生[16]和小麦[17]等作物均利用元分析对不同性状展开了研究,在水稻方面,前人利用元分析对产量性状[18]、干旱胁迫[19]相关的QTL作了报道,将控制性状遗传的位点进行了优化。然而利用元分析在水稻耐盐性QTL方面的研究尚未见报道。
因此,本研究收集了来自24个不同作图群体共344个在盐胁迫下鉴定的不同性状的QTL,利用McCouch Lab 2002、Cornell SSR 2001、Cornell IR64 × Azu DH QTL 2001、MU CT9993 × IR6226 QTL 2004、KRGRP 1998和IGCN 1998共6个图谱的整合图谱作为公共图谱,通过元分析获得一致性QTL,对区间小且具有多效性的一致性QTL,进行区间候选基因预测及功能分析,并对水稻基因数据库中的耐盐基因进行保守结构域分析,找到水稻耐盐功能候选基因,为水稻耐盐候选基因的发掘和分子标记辅助选择育种提供参考。
1 材料和方法
1.1 水稻耐盐相关性状QTL的收集
已报道的水稻耐盐性QTL从中国知网(http://epub.cnki.net/kns/default.htm)和NBCI(https://www.ncbi.nlm.nih.gov/pubmed)网站上收集和水稻耐盐性QTL研究的报道,统计QTL位置、置信区间、贡献率、加性效应、LOD值和连锁标记等信息。从Gramene网站下载6个水稻遗传图谱。
1.2 水稻耐盐相关性状QTL的信息整合及元分析
对收集的QTL信息按照BioMercator V4.2所需格式存为工作表,然后编辑为xlm格式的文件,文件包括图谱信息和QTL信息两方面。通过Genetic data loading将QTL所带图谱和6个参考图谱分别导入BioMercator V4.2,首先利用InforMap和ConsMap计算模块将6个图谱整合到一起,作为用于映射的图谱,再将QTL图谱和公共图谱进行整合,得到一致性图谱。然后利用QTL meta-analyses模块进行一致性QTL的挖掘,其原理为N个独立存在的与一个性状相关并且位于同一染色体同一位点附近的QTL计算出一个Meta-QTL。BioMercator V4.2软件对Meta-QTL的分析给出了5个模型的结果,每一个模型都按照最大似然函数比的方法通过高斯定理给出在染色体上最可能排列的位置和置信区间,由AIC最小值给出最优QTL模型即Meta-QTL[20]。
1.3 水稻耐盐性一致性QTL区间的物理定位及候选基因Gene ontology(GO)分析
利用http://rice.plantbiology.msu.edu/网站中的下载工具,下载基因组注释数据,将水稻物理图谱的序列信息与连锁图谱的标记信息进行整合,将一致性QTL两端的标记区间进行物理定位,同时下载一致性QTL区间的所有候选基因,利用WEGO(http://wego.genomics.org.cn/cgi-bin/wego/index.pl)在线分析软件进行Gene ontology 分析[21]。以了解水稻耐盐QTL区间所包含的生物学功能。
1.4 水稻耐盐性一致性QTL区间候选基因发掘
从水稻基因组数据库GRAMENE中查找在一致性QTL区间内已克隆的水稻和玉米耐盐基因,在NCBI上下载这些基因的蛋白质序列,并对其进行保守结构域分析,从而在一致性QTL区间内发掘水稻耐盐相关候选基因。
2 结果与分析
2.1 水稻耐盐性QTL信息收集
通过中国知网(http://epub.cnki.net/kns/default.htm)和NBCI(https://www.ncbi.nlm.nih.gov/pubmed)网站,共收集和水稻耐盐性QTL研究的报道18篇,共包含24张图谱,所用群体包括回交群体,重组自交系群体和F2群体等,计344个QTL。这些QTL涉及性状包括钠离子钾离子含量、死叶率、死苗率、苗存活天数、盐害级别、根苗性状及产量构成性状等方面(表1),6张水稻遗传公共图谱分别为McCouch Lab 2002[22]、Cornell SSR 2001[23]、Cornell IR64 × Azu DH QTL 2001[24]、MU CT9993 × IR6226 QTL 2004[25]、KRGRP 1998[26]和IGCN 1998[27]。
表1 水稻耐盐性QTL信息整合
Tab.1 Summary of the QTL of salt tolerance in rice reported previously
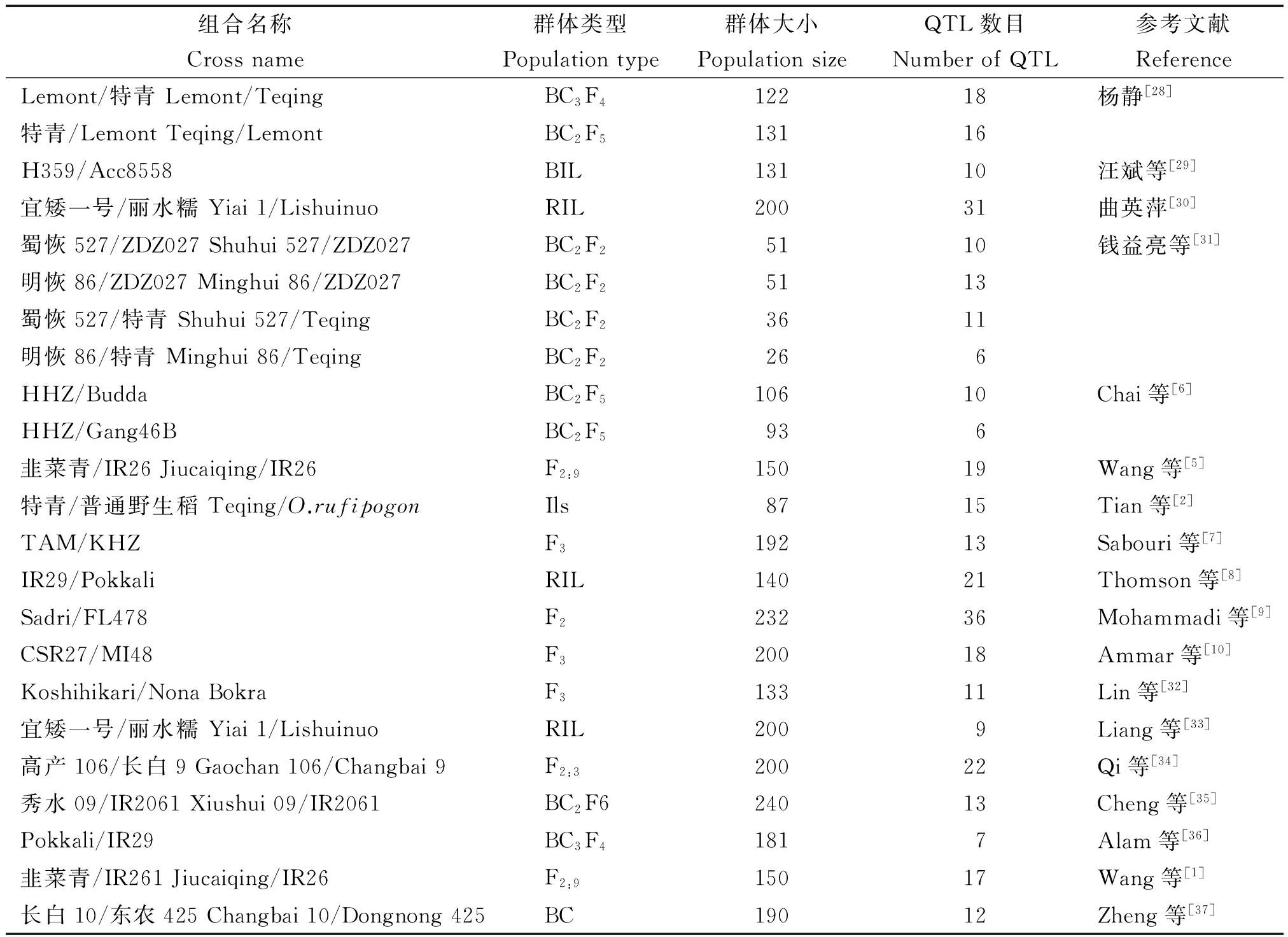
组合名称群体类型群体大小QTL数目参考文献CrossnamePopulationtypePopulationsizeNumberofQTLReferenceLemont/特青Lemont/TeqingBC3F412218杨静[28]特青/LemontTeqing/LemontBC2F513116H359/Acc8558BIL13110汪斌等[29]宜矮一号/丽水糯Yiai1/LishuinuoRIL20031曲英萍[30]蜀恢527/ZDZ027Shuhui527/ZDZ027BC2F25110钱益亮等[31]明恢86/ZDZ027Minghui86/ZDZ027BC2F25113蜀恢527/特青Shuhui527/TeqingBC2F23611明恢86/特青Minghui86/TeqingBC2F2266HHZ/BuddaBC2F510610Chai等[6]HHZ/Gang46BBC2F5936韭菜青/IR26Jiucaiqing/IR26F2:915019Wang等[5]特青/普通野生稻Teqing/O.rufipogonIls8715Tian等[2]TAM/KHZF319213Sabouri等[7]IR29/PokkaliRIL14021Thomson等[8]Sadri/FL478F223236Mohammadi等[9]CSR27/MI48F320018Ammar等[10]Koshihikari/NonaBokraF313311Lin等[32]宜矮一号/丽水糯Yiai1/LishuinuoRIL2009Liang等[33]高产106/长白9Gaochan106/Changbai9F2:320022Qi等[34]秀水09/IR2061Xiushui09/IR2061BC2F624013Cheng等[35]Pokkali/IR29BC3F41817Alam等[36]韭菜青/IR261Jiucaiqing/IR26F2:915017Wang等[1]长白10/东农425Changbai10/Dongnong425BC19012Zheng等[37]
2.2 水稻耐盐性QTL信息的图谱整合
通过对6张公共图谱进行整合,共有7 230个标记被整合到12条染色体上,对收集到的344个QTL进行映射,共有236个QTL被映射到水稻的12条染色体上(图1),各条染色体上QTL数分别为10,32,16,20,16,39,23,15,14,17,25,9个(表2)。
水稻耐盐相关性状QTL在图谱上存在成簇分布现象,如表2示,QTL数大于5个的Mqtl共有19个。在2号染色体的C2939~R2344、RM7288~RM145和R3041~RM240区间分别有8,16,7个QTL,3号染色体的OSR13~RM6496和C816~RM570区间分别富集了6,7个QTL,第4号染色体的R2373~RM335和C513~S12448区间分别集中了8,6个QTL,5号染色体的S12447~RM122区间包含5个QTL,6号染色体上的C916B~RM225、R2123~RM4924和RM3330~RM7179区间包含的QTL数分别为9,11,16个,7号染色体上的4个富集区包含的QTL数分别为6,5,5,7个,与之相连锁的区间为RM129~RG128、RM501~RM3859、RM11~RM5380、C451~RM560,8号染色体的RM8081~RM3572和RM4955~RM8271区间分别集中了5,7个QTL,10号染色体的RM5620~RM258区间和11号染色体的P97~RG211区间分别富集了8,16个QTL。这些QTL重叠群多数由水稻耐盐相关性状QTL组成。
2.3 水稻耐盐性QTL信息元分析
利用BioMercator V4.2中的QTL meta-analyses程序对整合的236个QTL进行元分析,以每次运算后AIC最小值为原则,最终得到46个一致性QTL。
被映射到水稻的12条染色体上的236个QTL包含在46个Mqtl中,其中初始QTL数大于3个的Mqtl共有39个,除了8号和12号染色体上均有3个Mqtl外,其余染色体上均有4个Mqtl,这些Mqtl的平均置信区间为2.21 cM。
在所有Mqtl中,发现了置信区间较小且包含多个性状的Mqtl。其中,位于1号染色体RM1232~RM104区间的0.67 cM遗传距离里,Mqtl1-2包含4个QTL,跟幼苗存活天数、苗高及穗长有关;Mqtl2-2和Mqtl2-3的置信区间分别为0.47,0.41 cM,分别和4,9个性状相关,包括钠钾离子浓度、盐害级别等性状;Mqtl4-2和Mqtl4-3位于P158~RM261和C513~S12448区间里,遗传距离分别为0.46,0.54 cM,另外,Mqtl6-3、Mqtl6-4、Mqtl7-3、Mqtl7-4和Mqtl10-4的置信区间均小于0.1 cM,与之相连锁的区间分别为R2123~RM4924、RM3330~RM7179、RM11~RM5380、C451~RM560和RM6469~RM6132。在46个Mqtl中,有37个Mqtl的遗传距离比初始QTL中置信区间最小的还小,说明元分析缩小了原始QTL的区间距离。

图1 水稻耐盐相关性状QTL元分析
Fig.1 meta-analysis of QTL related to salt tolerance traits in rice
表2 水稻耐盐性QTL元分析
Tab.2 The meta-analysis of QTL related to salt tolerance in rice
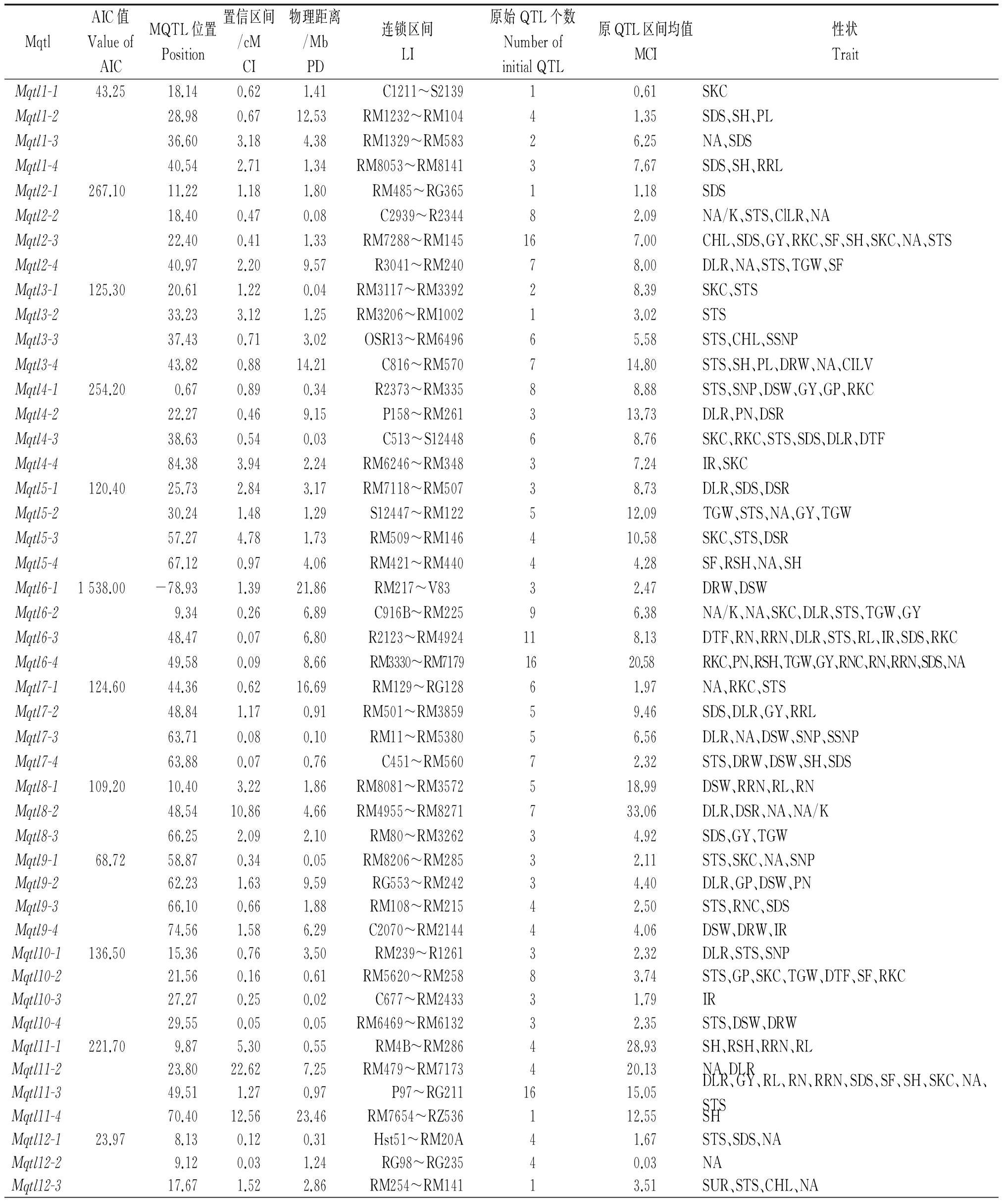
MqtlAIC值ValueofAICMQTL位置Position置信区间/cMCI物理距离/MbPD连锁区间LI原始QTL个数NumberofinitialQTL原QTL区间均值MCI性状TraitMqtl1-143.2518.140.621.41C1211~S213910.61SKCMqtl1-228.980.6712.53RM1232~RM10441.35SDS、SH、PLMqtl1-336.603.184.38RM1329~RM58326.25NA、SDSMqtl1-440.542.711.34RM8053~RM814137.67SDS、SH、RRLMqtl2-1267.1011.221.181.80RM485~RG36511.18SDSMqtl2-218.400.470.08C2939~R234482.09NA/K、STS、ClLR、NAMqtl2-322.400.411.33RM7288~RM145167.00CHL、SDS、GY、RKC、SF、SH、SKC、NA、STSMqtl2-440.972.209.57R3041~RM24078.00DLR、NA、STS、TGW、SFMqtl3-1125.3020.611.220.04RM3117~RM339228.39SKC、STSMqtl3-233.233.121.25RM3206~RM100213.02STSMqtl3-337.430.713.02OSR13~RM649665.58STS、CHL、SSNPMqtl3-443.820.8814.21C816~RM570714.80STS、SH、PL、DRW、NA、CILVMqtl4-1254.200.670.890.34R2373~RM33588.88STS、SNP、DSW、GY、GP、RKCMqtl4-222.270.469.15P158~RM261313.73DLR、PN、DSRMqtl4-338.630.540.03C513~S1244868.76SKC、RKC、STS、SDS、DLR、DTFMqtl4-484.383.942.24RM6246~RM34837.24IR、SKCMqtl5-1120.4025.732.843.17RM7118~RM50738.73DLR、SDS、DSRMqtl5-230.241.481.29S12447~RM122512.09TGW、STS、NA、GY、TGWMqtl5-357.274.781.73RM509~RM146410.58SKC、STS、DSRMqtl5-467.120.974.06RM421~RM44044.28SF、RSH、NA、SHMqtl6-11538.00-78.931.3921.86RM217~V83 32.47DRW、DSWMqtl6-29.340.266.89C916B~RM22596.38NA/K、NA、SKC、DLR、STS、TGW、GYMqtl6-348.470.076.80R2123~RM4924118.13DTF、RN、RRN、DLR、STS、RL、IR、SDS、RKCMqtl6-449.580.098.66RM3330~RM71791620.58RKC、PN、RSH、TGW、GY、RNC、RN、RRN、SDS、NAMqtl7-1124.6044.360.6216.69RM129~RG12861.97NA、RKC、STSMqtl7-248.841.170.91RM501~RM385959.46SDS、DLR、GY、RRLMqtl7-363.710.080.10RM11~RM538056.56DLR、NA、DSW、SNP、SSNPMqtl7-463.880.070.76C451~RM56072.32STS、DRW、DSW、SH、SDSMqtl8-1109.2010.403.221.86RM8081~RM3572518.99DSW、RRN、RL、RNMqtl8-248.5410.864.66RM4955~RM8271733.06DLR、DSR、NA、NA/KMqtl8-366.252.092.10RM80~RM326234.92SDS、GY、TGWMqtl9-168.7258.870.340.05RM8206~RM28532.11STS、SKC、NA、SNPMqtl9-262.231.639.59RG553~RM24234.40DLR、GP、DSW、PNMqtl9-366.100.661.88RM108~RM21542.50STS、RNC、SDSMqtl9-474.561.586.29C2070~RM214444.06DSW、DRW、IRMqtl10-1136.5015.360.763.50RM239~R126132.32DLR、STS、SNPMqtl10-221.560.160.61RM5620~RM25883.74STS、GP、SKC、TGW、DTF、SF、RKCMqtl10-327.270.250.02C677~RM243331.79IRMqtl10-429.550.050.05RM6469~RM613232.35STS、DSW、DRWMqtl11-1221.709.875.300.55RM4B~RM286428.93SH、RSH、RRN、RLMqtl11-223.8022.627.25RM479~RM7173420.13NA、DLRMqtl11-349.511.270.97P97~RG2111615.05DLR、GY、RL、RN、RRN、SDS、SF、SH、SKC、NA、STSMqtl11-470.4012.5623.46RM7654~RZ536112.55SHMqtl12-123.978.130.120.31Hst51~RM20A41.67STS、SDS、NAMqtl12-29.120.031.24RG98~RG23540.03NAMqtl12-317.671.522.86RM254~RM14113.51SUR、STS、CHL、NA
注:CI.置信区间;PD.物理距离;LI.连锁区间;MCI.平均置信区间;DTF.开花日期;SF.小穗育性;GY.谷物产量;RL.根长;DRW.根干质量;RKC.根部K+浓度;RN.根数;SKC.茎部K+浓度;DSW.杆干质量;SSNP.单株不育穗数;SNP.单株穗数;SUR.苗存活率;SDS.苗存活天数;SH.苗高;STS.盐害级别;NA/K.钠钾比;NA.钠离子;TGW.千粒质量;DSR.死苗率;DLR.死叶率;PL.穗长;PN.穗数;IR.吸收速率;RRL.相对根长;RRN.相对根数;RSH.相对苗高;CHL.叶绿素含量;CILV.营养生长期叶片Cl- 含量;CILR.生殖期叶片Cl- 含量。
Note:CI.Confidence interval;PD.Physical distance;LI.Linkage interval;MCI.Mean of confidence interval.DTF.Days to flowering;SF. Spikelet fertility;GY.Grain yield;RL.Root length;DRW.Dry root weight;RKC.Root K+ concentration;RN.Root number;SKC.Shoot K+ concentration;DSW.Dry steam weight;SSNP.Number of sterile spikelets per plant;SNP.Number of spikelets per plant;SUR.Seedling survival;SDS.Survival days of seedlings;SH.Seedling height;STS.Score of salt toxicity;NA/K.Na+/K+ ratio;NA.Na+;TGW.Thousand grains weight;DSR.Dead seedling rate;DLR.The dead leaf rate;PL.Panicle length;PN.Panicle number;IR.Imbibition rate;RRL.Relative root length;RRN.Relative root number;RSH.Relative seedling height;CHL.Leaf chlorophyll content;CILV.Cl- in leaf for vegetative;CILR.Cl- in leaf for reproductive.
2.4 水稻耐盐性一致性QTL区间的物理定位及候选基因Gene Ontology(GO)分析
对46个一致性QTL两端标记在水稻物理图谱RIGSP2005上的位置进行物理定位,结果表明,这46个一致性QTL的平均物理距离为4.41 Mb,其中物理距离小于1 Mb有13个(表2),这13个一致性QTL中,有8个遗传距离在0.5 cM附近(表3),且富集多个QTL。这8个一致性QTL范围内共包含1 489个预测基因,Mqtl12-1区间基因数最多(724个),Mqtl10-4区间内基因数最少(22个),其中Mqtl7-4在染色体上的覆盖范围最大(0.763 Mb),Mqtl4-3的覆盖范围最小(0.033 Mb)。
利用Wego在线分析软件对8个一致性QTL区间的1 489个预测基因进行GO分析,结果表明,有364个基因参与了细胞组成成分(图2-A),470个基因参与了分子功能(图2-B),669个基因参与了生物进程(图2-C),分别涉及19种细胞成分、16种分子功能和21种生物进程。其中在细胞组成成分中,细胞、细胞合成成分、细胞质、细胞溶质、细胞膜、原生质膜和质体所占比例较大,在分子功能中,结合物、催化活性、水解酶活性、分子功能和蛋白结合相关的基因所占比例较大,生物进程中,生物学过程、细胞形成和新陈代谢所占比例较大。其中有18个被注释为与非生物胁迫相关的基因。这些基因分别与脂氧合酶(LOC_Os02g10120)、玉米黄素环氧酶(LOC_Os04g37619)、Dof基因家族转录因子(LOC_Os07g32510)、热激蛋白区域(LOC_Os07g33350)、谷氨酸受体(LOC_Os07g33790)、载体家族蛋白(LOC_Os07g33910)、多肽转运蛋白(LOC_Os10g33210)、花粉发育调控(LOC_Os10g33250)等功能相关。
2.5 水稻耐盐性一致性QTL区间候选基因发掘
通过GRAMENE数据库查找,共有39个耐盐性基因包含在一致性QTL区间中,同源比对后共发现9个和水稻耐盐相关的候选基因(表4),涉及BZIP转录因子、水解酶、锌指蛋白、DNA结构域、蛋白激酶等功能。其中Mqtl1-2区间内基因OsABI5和GRMZM2G138861在该区段内与OS01G0813100和OS01G0869900同源,序列相似度分别为79.2%和76.1%,Mqtl2-4区间内的OsTPP1基因在该区段内与OS02G0753000同源,序列相似度为67.3%,3号染色体的Mqtl3-3和Mqtl3-4上的rcn1和DST基因分别与对应区段内同源的基因是OS03G0282100和OS03G0607700,相似度分别为71.1%和70.00%,Mqtl7-1区间的OsCIPK23与OS07G0150500同源,序列完全相同,Mqtl9-2区间的OsDREB6和OsEATB分别与OS09G0369050和OS09G0434500同源,序列匹配度为100.0%和72.5%,Mqtl9-3区间内的OsDREB1B与OS09G0522100同源,序列匹配度为85.0%。
表3 Mqtl区间的物理图谱定位
Tab.3 The Mqtl region on rice physical map of IRGSP2005
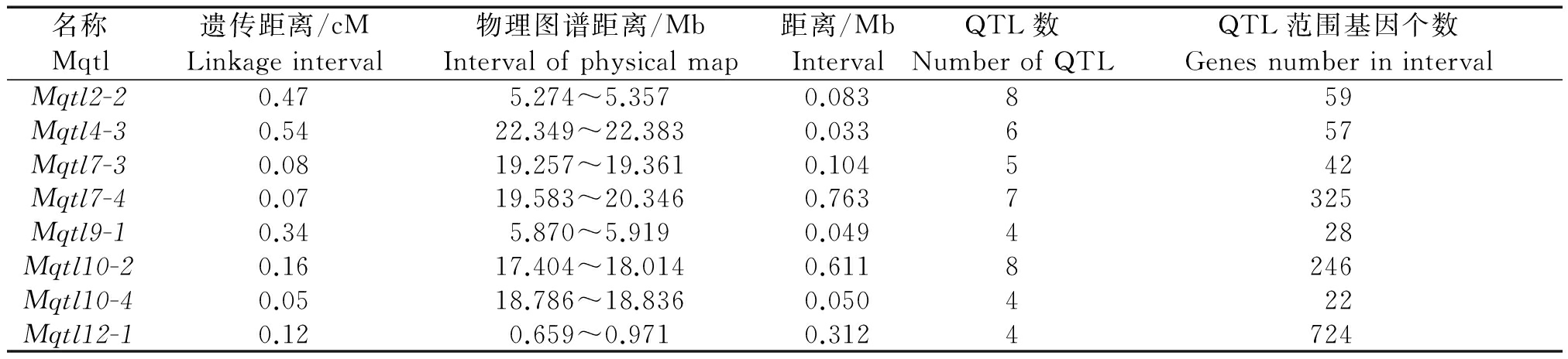
名称遗传距离/cM物理图谱距离/Mb距离/MbQTL数QTL范围基因个数MqtlLinkageintervalIntervalofphysicalmapIntervalNumberofQTLGenesnumberinintervalMqtl2-20.475.274~5.3570.083859Mqtl4-30.5422.349~22.3830.033657Mqtl7-30.0819.257~19.3610.104542Mqtl7-40.0719.583~20.3460.7637325Mqtl9-10.345.870~5.9190.049428Mqtl10-20.1617.404~18.0140.6118246Mqtl10-40.0518.786~18.8360.050422Mqtl12-10.120.659~0.9710.3124724

图2 一致性QTL区间候选基因GO分析
Fig.2 Gene Ontology analysis of genes related to consensus QTL
表4 Mqtl区间水稻耐盐基因与水稻基因组预测基因序列的一致性
Tab.4 Coherence of gene loci and predicted genes from rice sequence in Mqtl region
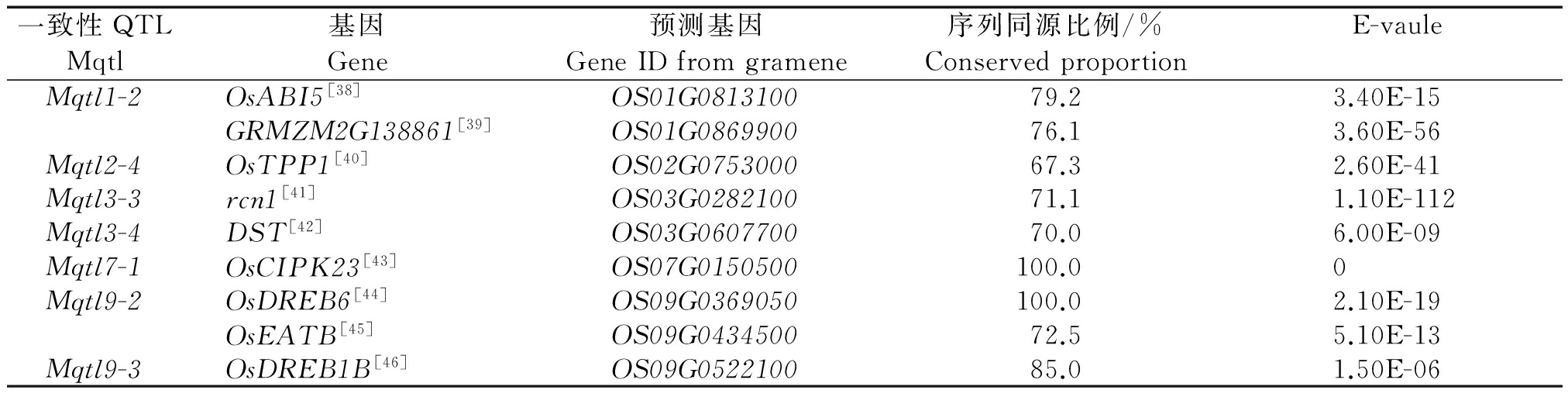
一致性QTL基因预测基因序列同源比例/%E-vauleMqtlGeneGeneIDfromgrameneConservedproportionMqtl1-2OsABI5[38]OS01G081310079.23.40E-15GRMZM2G138861[39]OS01G086990076.13.60E-56Mqtl2-4OsTPP1[40]OS02G075300067.32.60E-41Mqtl3-3rcn1[41]OS03G028210071.11.10E-112Mqtl3-4DST[42]OS03G060770070.06.00E-09Mqtl7-1OsCIPK23[43]OS07G0150500100.00Mqtl9-2OsDREB6[44]OS09G0369050100.02.10E-19OsEATB[45]OS09G043450072.55.10E-13Mqtl9-3OsDREB1B[46]OS09G052210085.01.50E-06
3 讨论与结论
利用现存的生物信息学工具可以获得任何QTL区间内的基因[47],研究利用的公共遗传图谱具有7 230个分子标记,它提供了丰富的图谱标记,对水稻耐盐相关性状的344个QTL进行整理,共有236个被整合到水稻6个遗传公共图谱上,12条染色体上均有分布,可见水稻耐盐性是复杂的数量性状,受多基因控制。Swamy等[18]对水稻产量性状进行了元分析,将产量性状QTL最小界定到500 kb以内,Khowaja等[48]对水稻耐旱性QTL进行了元分析,在一致性QTL区间找到了180个候选基因,Mideros等[49]对玉米霉菌毒素的QTL元分析后发现了一个贡献率较大的一致性QTL,这些研究为MAS提供了很好参考。本研究中,在检测到的一致性QTL中,有19个区间的QTL数大于5个,存在成簇分布现象,且有37个Mqtl的遗传距离比初始QTL中置信区间最小的还小,说明元分析缩小了原始QTL的区间距离。检测到的46个一致性QTL的平均物理距离为4.41 Mb,其中物理距离小于1 Mb有13个,这13个一致性QTL中,有8个的遗传距离在0.5 cM附近,且富集多个QTL。这8个一致性QTL范围内共包含1 489个预测基因,其中Mqtl10-4区间内基因数最少(22个),Mqtl4-3的覆盖范围最小(0.033 Mb)。GO分析发现,在1 489个预测基因中,绝大多数参与了细胞合成、结合功能、催化活性、水解酶活性、分子作用和蛋白结合相关生物过程,并找到了18个与非生物胁迫相关的基因,可能与盐胁迫表达相关联。可见利用高密度的遗传图谱进行QTL整合及元分析能高效的将控制性状表达的QTL精细化,并预测区段内基因的功能,是开展水稻耐盐MAS和基因克隆的有效途径之一。
为了分析一致性QTL区间内和耐盐基因蛋白序列相同或相似的预测基因,对46个一致性QTL区间内的39个与盐胁迫相关的基因进行了蛋白序列比对,共有8个水稻耐盐基因找到了预测基因,另外,由于禾本科作物的基因组存在高度的保守性,所以某一作物的QTL或基因信息可以用于其他作物[50],因此,本研究对玉米耐盐基因进行了同源比对,在Mqtl1-2区间仅发掘了1个与GRMZM2G138861同源的水稻耐盐预测基因,可能是由于玉米基因组比水稻要大,遗传转化没有水稻研究成熟,导致玉米耐盐基因研究缓慢。这9个预测基因和与之对应的耐盐基因具有高度的蛋白序列相似性,且保守结构域高度一致,可用于开展候选基因验证,利用qRT-PCR技术确定候选基因表达模式,并在关联群体中对性状进行候选基因关联分析,为进一步克隆水稻耐盐基因奠定基础。
研究发现了5个遗传距离小于0.1 cM,且具有多效性的Mqtl,两端的分子标记可用于分子标记辅助育种。在发现的8个物理距离在0.5 Mb附近的Mqtl中,找到了9个和水稻耐盐基因具有高度蛋白序列的相似性的注释基因,作为水稻耐盐的候选基因。分别为OS01G0813100、OS01G0869900、OS02G0753000、OS03G0282100、OS03G0607700、OS07G0150500、OS09G0369050、OS09G0434500和OS09G0522100。
参考文献:
[1] Wang Z,Chen Z,Cheng J,et al.QTL analysis of Na+ and K+ concentrations in roots and shoots under different levels of NaCl stress in rice (Oryza sativa L.)[J].PloS One,2012,7 (12):e51202.
[2] Tian L,Tan L B,Liu F X,et al.Identification of quantitative trait loci associated with salt tolerance at seedling stage from Oryza rufipogon[J].Journal of Genetics and Genomics,2011,38(12):593-601.
[3] Ren Z,Chao D.QTLs for Na+and K+uptake of the shoots and roots controlling rice salt tolerance[J].Theoretical and Applied Genetics,2004,108(2):253-260.
[4] Prasad S,Bagali P,Hittalmani S,et al.Molecular mapping of quantitative trait loci associated with seedling tolerance to salt stress in rice (Oryza sativa L.)[J].Current Science,2000,78 (2):162-164.
[5] Wang Z F,Wang J F,Bao Y M,et al.Quantitative trait loci controlling rice seed germination under salt stress[J].Euphytica,2011,178(3):297-307.
[6] Chai L,Zhang J,Pan X B,et al.Advanced backcross QTL analysis for the whole plant growth duration salt tolerance in rice (Oryza sativa L.)[J].Journal of Integrative Agriculture,2014,13(8):1609-1620.
[7] Sabouri H,Rezai A,Moumeni A,et al.QTLs mapping of physiological traits related to salt tolerance in young rice seedlings[J].Biologia Plantarum,2009,53(4):657-662.
[8] Thomson M J,De Ocampo M,Egdane J A,et al.Characterizing the saltol quantitative trait locus for salinity tolerance in rice[J].Rice,2010,3(2/3):148-160.
[9] Mohammadi R,Mendioro M S,Diaz G Q,et al.Mapping quantitative trait loci associated with yield and yield components under reproductive stage salinity stress in rice(Oryza sativa L.)[J].Journal of Genetics,2013,92(3):433-443.
[10] Ammar M H M,Pandit A,Singh R K,et al.Mapping of QTLs controlling Na+,K+ and Cl- ion concentrations in salt tolerant indica rice variety CSR27[J].Journal of Plant Biochemistry and Biotechnology,2009,18(2):139-150.
[11] Deshmukh R K,Sonah H,Kondawar V,et al.Identification of meta quantitative trait loci for agronomical traits in rice (Oryza sativa)[J].Indian Journal of Genetics and Plant Breeding,2012,72 (3):264-270
[12] Arcade A,Labourdette A,Falque M,et al.BioMercator:integrating genetic maps and QTL towards discovery of candidate genes[J].Bioinformatics,2004,20(14):2324-2326.
[13] Zhang J F,Yu J W,Pei W F,et al.Genetic analysis of Verticillium wilt resistance in a backcross inbred line population and a meta-analysis of quantitative trait loci for disease resistance in cotton[J].BMC Genomics,2015,16(1):577.
[14] Shinozuka H,Cogan N O,Spangenberg G C,et al.Quantitative trait locus (QTL) meta-analysis and comparative genomics for candidate gene prediction in perennial ryegrass (Lolium perenne L.)[J].BMC genetics,2012,13 (1):101.
[15] Zhang H,Uddin M S,Zou C,et al.Meta-analysis and candidate gene mining of low-phosphorus tolerance in maize[J].Journal of Integrative Plant Biology,2014,56(3):262-270.
[16] Dong Y B,Zhang Z W,Shi Q L,et al.QTL identification and meta-analysis for kernel composition traits across three generations in popcorn[J].Euphytica,2015,204(3):649-660.
[17] Tyagi S,Mir R,Balyan H,et al.Interval mapping and meta-QTL analysis of grain traits in common wheat (Triticum aestivum L.)[J].Euphytica,2015,201 (3):367-380.
[18] Swamy B ,Sarla N.Meta-analysis of Yield QTLs derived from inter-specific crosses of rice reveals consensus regions and candidate genes[J].Plant Molecular Biology Reporter,2011,29(3):663-680.
[19] Trijatmiko K R,Supriyanta,Prasetiyono J A,et al.Meta-analysis of quantitative trait loci for grain yield and component traits under reproductive-stage drought stress in an upland rice population[J].Molecular Breeding,2014,34(2):283-295.
[20] Goffinet B,Gerber S.Quantitative trait loci:a meta-analysis[J].Genetics,2000,155(1):463-473.
[21] Ashburner M,Ball C A,Blake J A,et al.Gene ontology:tool for the unification of biology[J].Nature Genetics,2000,25(1):25-29.
[22] Mccouch S R,Teytelman L,Xu Y,et al.Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.)[J]. DNA research,2002,9 (6):199-207.
[23] Temnykh S D G.Experimental analysis of microsatellites in rice.(Oryza sativa L.):frequency,length variation,transposon associations,and genetic marker potential[J].Genome Research,2001,11(8):1441-1452.
[24] Septiningsih E M,Prasetiyono J,Lubis E,et al.Identification of quantitative trait loci for yield and yield components in an advanced backcross population derived from the Oryza sativa variety IR64 and the wild relative O.rufipogon[J].Theoretical and Applied Genetics,2003,107(8):1419-1432.
[25] Babu R C,Nguyen B D,Chamarerk V,et al.Genetic analysis of drought resistance in rice by molecular markers:Association between secondary traits and field performance[J].Crop Science,2003,43(4):1457-1469.
[26] Cho Y G,McCouch S R,Kuiper M,et al.Integrated map of AFLP,SSLP and RFLP markers using a recombinant inbred population of rice (Oryza sativa L.)[J].Theoretical and Applied Genetics,1998,97 (3):370-380.
[27] Harushima Y,Yano M,Shomura P,et al.A high-density rice genetic linkage map with 2275 markers using a single F-2 population[J].Genetics,1998,148(1):479-494.
[28] 杨 静.利用双向导入系剖析水稻耐盐QTL定位的遗传背景效应[D].哈尔滨:东北农业大学,2009.
[29] 汪 斌,兰 涛,吴为人.盐胁迫下水稻苗期Na+含量的QTL定位[J].中国水稻科学,2007,21(6):585-590.
[30] 曲英萍.水稻耐盐碱性QTLs分析[M].北京:中国农业科学院,2007.
[31] 钱益亮,王 辉,陈满元,等.利用BC2F3产量选择导入系定位水稻耐盐QTL[J].分子植物育种,2009,7(2):224-232.
[32] Lin H X,Zhu M Z,Yano M,et al.QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance[J].Theoretical and Applied Genetics 2004,108(2):253-260.
[33] Liang J L,Qu Y P,Yang C G,et al.Identification of QTLs associated with salt or alkaline tolerance at the seedling stage in rice under salt or alkaline stress[J].Euphytica,2015,201(3):441-452.
[34] Qi D,Guo G,Lee M C,et al.Identification of quantitative trait loci for the dead leaf rate and the seedling dead rate under alkaline stress in rice[J].Journal of Genetics and Genomics,2008,35(5):299-305.
[35] Cheng L R,Wang Y,Meng L J,et al.Identification of salt-tolerant QTLs with strong genetic background effect using two sets of reciprocal introgression lines in rice[J].Genome,2012,55(1):45-55.
[36] Alam R,Sazzadur R M,Seraj Z I,et al.Investigation of seedling-stage salinity tolerance QTLs using backcross lines derived from Oryza sativa L.[J].Plant Breeding,2011,130(4):430-437.
[37] Zheng H L,Zhao H W,Liu H L,et al.QTL analysis of Na+ and K+ concentrations in shoots and roots under NaCl stress based on linkage and association analysis in japonica rice[J].Euphytica,2015,201(1):109-121.
[38] Zou M,Guan Y,Ren H,et al.A bZIP transcription factor,OsABI5,is involved in rice fertility and stress tolerance[J].Plant Molecular Biology,2008,66(6):675-683.
[39] Ying S,Zhang D F,Li H Y,et al.Cloning and characterization of a maize SnRK2 protein kinase gene confers enhanced salt tolerance in transgenic Arabidopsis[J].Plant Cell Reports, 2011,30 (9):1683-1699.
[40] Pramanik M,Imai R.Functional identification of a trehalose 6-phosphate phosphatase gene that is involved in transient induction of trehalose biosynthesis during chilling stress in rice[J].Plant Molecular Biology,2005,58(6):751-762.
[41] Matsuda S,Nagasawa H,Yamashiro N A,et al.Rice RCN1/OsABCG5 mutation alters accumulation of essential and nonessential minerals and causes a high Na/K ratio,resulting in a salt-sensitive phenotype[J].Plant Science,2014,224(9):103-111.
[42] Huang X Y,Chao D Y,Gao J P,et al.A previously unknown zinc finger protein,DST,regulates drought and salt tolerance in rice via stomatal aperture control[J].Genes & Development,2009,23(15):1805-1817.
[43] Li J,Long Y,Qi G N,et al.The Os-AKT1 channel is critical for K+ uptake in rice roots and is modulated by the rice CBL1-CIPK23 complex[J].The Plant Cell,2014,26(8):3387-3402.
[44] Ke Y G,Yang Z J,Yu S W,et al.Characterization of OsDREB6 responsive to osmotic and cold stresses in rice[J].Journal of Plant Biology,2014,57(3):150-161.
[45] Qi W,Sun F,Wang Q,et al.Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene[J].Plant Physiology,2011,157(1):216-228.
[46] Dubouzet J G, Sakuma Y, Ito Y, et al. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-,high-salt-and cold-responsive gene expression[J]. The Plant Journal, 2003, 33(4): 751-763.
[47] Droc G,Ruiz M,Larmande P,et al.OryGenesDB:a database for rice reverse genetics[J].Nucleic Acids Research,2006,34:736-740.
[48] Khowaja F S,Gj N,Courtois B,et al.Improved resolution in the position of drought-related QTLs in a single mapping population of rice by meta-analysis[J].BMC Genomics,2009,10(1):276.
[49] Mideros S X,Warburton M L,Jamann T M,et al.Quantitative trait loci influencing mycotoxin contamination of maize:analysis by linkage mapping,characterization of Near-Isogenic lines,and Meta-Analysis[J].Crop Science,2014,54(1):127-142.
[50] Chardon F,Virlon B,Moreau L,et al.Genetic architecture of flowering time in maize as inferred from quantitative trait loci meta-analysis and synteny conservation with the rice genome[J].Genetics,2004,168(4):2169-2185.